Abstract
The complete mitochondrial genome (mitogenome) of the Kyrghyz racerunner (Eremias nikoskii Bedriaga, 1905) from Kyrgyzstan was determined for the first time by next-generation sequencing. The mitogenome was 20,840 bp in length and comprised the standard set of 13 protein-coding genes (PCGs), 2 ribosomal RNA genes, 22 transfer RNA genes, and a control region. The 13 concatenated PCGs were used to implement Bayesian phylogenetic analyses together with some congeners and three representative lacertids retrieved from GenBank. The monophyly of both Eremias and its viviparous group was recovered in the Bayesian phylogenetic tree, while the subgenus Pareremias was paraphyletic with respect to E. nikoskii. The mitogenome of E. nikoskii will faciliate the research on species delimitation, molecular evolution, and phylogenetic inference in the racerunner lizards.
The Kyrghyz racerunner, Eremias nikoskii Bedriaga, 1905, is an oviparous species among subgenus Aspidorhinus (Barabanov Citation2009) in genus Eremias. This species occurs in the mountain ranges around the Fergana valley in eastern Uzbekistan, Kyrgyzstan, and northern Tajikistan, and prefers mountain ravines and the valleys of mountain rivers, in rocky areas with thin grass and bush cover up to about 1000–3000 m above sea level (Szczerbak Citation2003). To date, little is known about its genetic affinities with congeners albeit with limited understanding of phylogeny of the racerunner (Eremias) lizards (Guo et al. Citation2011; Khan et al. Citation2021).
In this study, we reported the whole mitogenome of E. nikolskii for the first time, with voucher number Guo4717. This specimen was collected from westward of Kazarman village (41.38750˚N, 73.93999˚E, 1332 meters above sea level), Dzhalalabad region in Kyrgyzstan on 28 August 2014. Its liver tissue was dissected from the euthanized lizard, fixed with 95% ethanol, and stored at −20 °C in the Chengdu Institute of Biology, Chinese Academy of Sciences (contact person: Xianguang Guo, E-mail: [email protected]). The CIB Animal Care and Use Committee approved all procedures.
Total genomic DNA was extracted from the liver tissue in the Genepioneer Biotechnologies Co. Ltd. (Nanjing, China) for 150 bp paired-end (PE150) library construction as well as sequencing through the Illumina NovaSeq (Illumina, USA). The raw data were processed with fastp v0.20.0 (Chen et al. Citation2018) by trimming adapters and primers, and filtering low quality reads. Assembly of clean data was performed using SPAdes v3.10.1 (Bankevich et al. Citation2012). Subsequently, we took a similar strategy to that in Wang et al. (Citation2021) to obtain the complete mitogenome. The mitogenome was preliminarily annotated with MITOS Web Server (http://mitos2.bioinf.uni-leipzig.de; Bernt et al. Citation2013). Twenty-two tRNA genes were confirmed by using the software tRNA scan-SE (Lowe and Chan Citation2016). The nucleotide composition was estimated in MEGA v7.0 (Kumar et al. Citation2016).
The complete mitogenome of E. nikoskii was 20,840 bp in length, which was composed of 28.00% T, 28.01% C, 30.54% A, 13.45% G. A total of 37 genes were obtained including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNAs), 2 ribosomal RNA gens, and a control region (CR or D-loop). The gene content, order, and composition exhibited a typical vertebrate mtDNA feature. Most genes were distributed on H-strand, with exception to ND6 gene and eight tRNAs (tRNA-Glu, Ala, Asn, Cys, Tyr, Ser[UGA], Gln, and Pro). In the 13 PCGs, the shortest one was ATP8 gene (162 bp) and the longest one was ND5 (1824 bp). Only COX1 gene used GTG as a start codon, while the other PCGs used ATG. Five PCGs (ND1, ATP8, ATP6, ND4L, ND5) used TAA as stop codon; five PCGs (ND2, COX2, COX3, ND3, ND4) used T; two PCGs (COX1, ND6) used AGG; Cytb used TAG. In addition, 12S rRNA, 16S rRNA, and CR were 951, 1545, and 5436 bp, respectively.
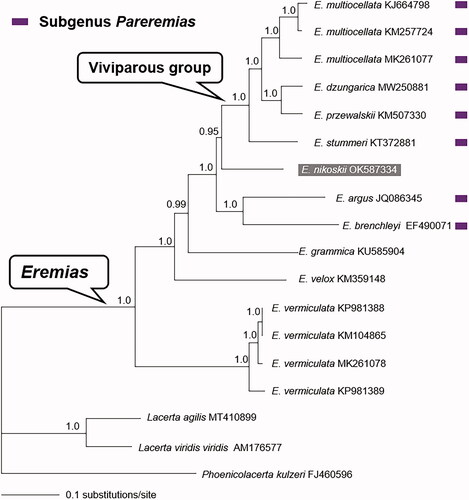
Phylogenetic trees were inferred from the concatenated PCGs of Eremias spp. and other representative lacertids (Lacertidae) retrieved from GenBank. Bayesian analyses were conducted using MrBayes v3.2.7a (Ronquist et al. Citation2012) with the GTR + G + I substitution model. A 50% majority-rule consensus tree was assessed by combining the sampled trees from two independent runs after a 30% burn-in phase. Clade support was assessed by posterior probability (PP). As shown in , the monophyly of both genus Eremias and its viviparous group was recovered with strong support, which is consistent with previous studies (Guo et al. Citation2011; Orlova et al. Citation2017; Liu et al. Citation2021). Eremias nikoskii was inferred as the sister taxon to the viviparous group with strong support (PP = 0.95); this challenged monophyly of the subgenus Pareremias (Arnold Citation1986; Guo et al. Citation2011). The mitogenome of E. nikoskii will facilitate the research on species delimitation, molecular evolution, and phylogenetic inference in the racerunner lizards.
Author contributions
Conceptualization, X.G.; Data curation, J.L. and X.G.; Analysis and interpretation of the data, X.G., X.H. and J.L.; Funding acquisition, J.L. and X.G.; Writing – original draft, X.G. and X.H.; Writing – review & editing, X.G., X.H., J.L. and M.C. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
We thank Dr. Roman A. Nazarov (Zoological Museum, Department of Herpetology, Moscow State University) for assistance in collecting the sample during the fieldwork. We are grateful to Mr. Song Wang for assistance in submitting the unassembled, high throughput sequencing reads to SRA of NCBI.
Disclosure statement
No potential conflict of interest was reported by the author(s). The authors alone are responsible for the content and writing of this article.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/nuccore/OK587334) under the accession number OK587334. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA773219, SRR16509497, and SAMN22448698, respectively.
Additional information
Funding
References
- Arnold EN. 1986. The hemipenis of lacertid lizards (Reptilia: Lacertidae): structure, variation and systematic implications. J Natl Hist. 20(5):983–1257.
- Bankevich A, Nurk S, Antipov D, Gurevich A, Dvorkin M, Kulikov AS, Lesin V, Nikolenko S, Pham S, Prjibelski A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Barabanov AV. 2009. Aspidorhinus Eichwald, 1841 as a valid subgeneric name for Eremias velox species group (Sauria, Lacertidae). Curr Stud Herpetol. 9:59–61 (in Russian with English abstract).
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one fastq preprocessor. Bioinformatics. 34(17):i884–i890.
- Guo X, Dai X, Chen D, Papenfuss TJ, Ananjeva NB, Melnikov DA, Wang Y. 2011. Phylogeny and divergence times of some racerunner lizards (Lacertidae: Eremias) inferred from mitochondrial 16S rRNA gene segments. Mol Phylogenet Evol. 61(2):400–412.
- Khan MA, Jablonski D, Nadeem MS, Masroor R, Kehlmaier C, Spitzweg C, Fritz U. 2021. Molecular phylogeny of Eremias spp. from Pakistan contributes to a better understanding of the diversity of racerunners. J Zool Syst Evol Res. 59(2):466–483.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liu J-L, Dujsebayeva TN, Chirikova MA, Gong X, Li D-J, Guo X-G. 2021. Does the Dzungarian racerunner (Eremias dzungarica Orlova, Poyarkov, Chirikova, Nazarov, Munkhbaatar, Munkhbayar & Terbish, 2017) occur in China? Species delimitation and identification with DNA barcoding and morphometric analyses. Zool Res. 42(3):287–293.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Orlova VF, Poyarkov NA, Chirikova MA, Nazarov RA, Munkhbaatar M, Munkhbayar K, Terbish K. 2017. MtDNA differentiation and taxonomy of central asian racerunners of Eremias multiocellata-E. przewalskii species complex (Squamata, Lacertidae). Zootaxa. 4282(1):1–42.
- Ronquist F, Teslenko M, Mark PVD, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Szczerbak NN. 2003. Guide to the reptiles of the Eastern Palearctic. Malabar, Florida: Krieger Publishing Company.
- Wang S, Liu J, Zhang B, Guo X. 2021. The complete mitochondrial genome of Eremias dzungarica (Reptilia, Squamata, Lacertidae) from the Junggar Basin in Northwest China. Mitochondrial DNA B Resour. 6(7):2012–2014.