Abstract
Nyctocalos is a genus of flowering lianas belonging to the family Bignoniaceae, and occurring from South-Central China to Malesia. In this study, we assembled the first complete chloroplast genome of N. pinnatum. The total length of the chloroplast genome is 159591 bp, with a GC content of 38.04%, which includes a pair of inverted repeats of 30,480 bp, a small single-copy region of 12,774 bp and a large single-copy region of 85,857 bp. The chloroplast genome contains 135 genes, consisting of 89 protein-coding genes, 38 transfer RNAs, and 8 ribosomal RNAs. We constructed a phylogenomic tree with representative chloroplast genomes from Bignoniaceae. N. pinnatum is revealed to be sister to Oroxylum in the tribe Oroxyleae, with a high bootstrap support. This is the first chloroplast genome assembled in Nyctocalos, and it provides essential information for further ecology and evolutionary studies in this genus and Bignoniaceae.
Nyctocalos pinnatum Steenis 1953 is classified in Bignoniaceae, tribe Oroxyleae (Kubitzki Citation2004). The genus is characterized by its septicidal capsule and Asian distribution. Nyctocalos has three species native to South central China to West Malesia (van Steenis Citation1977; Kubitzki Citation2004). Two species are found in China, N. brunfelsiiflorum and N. pinnatum, although the classification and nomenclature of the genus is complex and the two species are often misidentified. Nyctocalos flowers have a long, tubular, white reflective and fragrant corollas that only open at night and are said to be pollinated by bats (Corlett Citation2004). As in other Bignoniaceae its stigmas are sensitive and will shut when touched by a visitor thus avoiding repetitive fertilization (Darwin Citation1876; Milet-Pinheiro et al. Citation2009). The Chinese name for this genus means shed light into the night, which reflects well in its ecology and morphology. Other morphological unique characters in Nyctocalos as well as Hieris are the punctate glands on leaves, calyx, and corolla which attract many ants.
Nyctocalos is placed under the tribe Oroxyleae, which includes three other genera, Millingtonia, Oroxylum, and Hieris. The phylogeny of the neotropical members of the Bignoniaceae family has been well studied based on two chloroplast markers, rbcL and ndhF (Spangler and Olmstead Citation1999), as well as an additional marker, trnL-F, and increased sampling (Olmstead et al. Citation2009), however, the phylogeny of Asian members has not been studied. The placement of tribe Oroxyleae is unresolved and only cladistic analyses of morphological traits is available (Abdel-Hameed Citation2014).
Here, we assembled and annotated the chloroplast genome of N. pinnatum using next-generation sequencing, in order to provide an opportunity for finer scale molecular and taxonomic work in this family and this tribe, such as providing super-barcodes for identification against closely related species (Chen et al. Citation2020).
We sampled fresh leaves of wild sourced N. pinnatum, collected in Yuxi, Yunnan, China, and cultivated in the vine garden of Xishuangbanna Tropical Botanical Garden (Yunnan, China, geospatial coordinates: 21°55′N 101°15′E; altitude: 570 m), Chinese Academy of Sciences. The collection permit was granted by the Center for Gardening and Horticulture of Xishuangbanna Tropical Botanical Garden. The herbarium specimen voucher was deposited in HITBC, Yunnan, China (contact info: Jianwu Li, [email protected]) under the voucher number V000030. Total genomic DNA was extracted using Magnetic Plant Genomic DNA Kit (TIANGEN). Ilumina NovaSeq 6000 platform was used for sequencing and pair-end reads with 150 bp each were generated. Libraries preparation and sequencing were performed by Annoroad Gene Technology (Beijing China).
Adaptor sequences were removed from raw data and low quality reads were filtered out. Clean data are used for de novo assembly of the chloroplast genome using GetOrganelle 1.7.5 (Jin et al. Citation2020). We then mapped the clean reads back to the assembled chloroplast genome to check for assembly errors. As for annotation, we manually compared the automated version generated from CpGAVAS2 (Shi et al. Citation2019) to Oroxylum indicum (NC_049086.1), which is a closely related species using Geneious (v2020.0.5). Finally, the chloroplast DNA sequence and annotations were deposited in GenBank under accession number OK649927.
The chloroplast genome is 159,591 bp in length, including the Large Single Copy (LSC) region (85,857 bp), the Small Single Copy (SSC) region (12,774 bp), and two Inverted Repeats (IR, 30,480 bp). The overall GC content is 38.04%, with the number for LSC and SSC being 36.32% and 33.51%, respectively, which are both lower than in the IR regions (41.4%). The chloroplast genome has 135 genes, consisting of 89 protein-coding, 38 tRNA, and 8 rRNA genes.
To further confirm the placement of Nyctocalos in Bignoniaceae, protein-coding sequences from complete chloroplast genomes of 27 Bignoniaceae species that are publicly available including N. pinnatum were used to reconstruct the phylogeny of this family. Sequence alignment was performed using MAFFT (Katoh and Standley Citation2013), topology and supporting rate of the phylogenetic tree were computed using RAxML v8.2.10 (Stamatakis Citation2014) using the GTR + G substitution model and with 1000 bootstrap. Our tree supports the placement of Nyctocalos as sister to Oroxylum, which is another genus in Oroxyleae (). The backbone relationship between major tribes largely agrees with Olmstead Citation2009, except that Oroxyleae is a sister group to Crescentiina (bootstrap = 83%) instead of Catalpeae (bootstrap = 48% as reported in Olmstead Citation2009).
Figure 1. Maximum-likelihood tree of N. pinnatum in relation to other 26 species in Bignoniaceae based on protein-coding sequences from complete chloroplast genomes. Bootstrap values are shown next to the nodes.
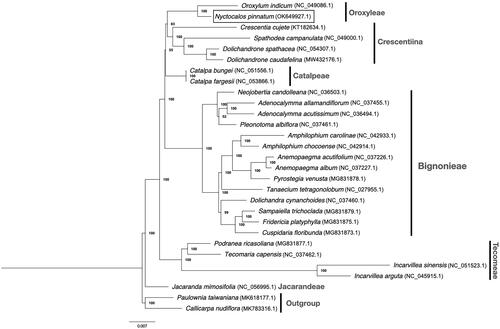
To conclude, the complete chloroplast genome of N. pinnatum is a useful DNA data source for further studies of the evolutionary history of this genus and tribe Oroxyleae and can be used as super-barcodes for identification of this species in ecological studies.
Author contributions
HF and SL were involved in the conception and design; HF and JL did the analysis; HF drafted the paper; SL revised it critically for intellectual content; all authors approved the final version to be published and agree to be accountable for all aspects of the work.
Acknowledgement
This work was supported by the Core Botanical Gardens under Grant No. Y9ZK011B08. HF was supported by the China Postdoctoral Fund (2019M663597) and International Postdoctoral Exchange Fellowship Program. We als would like to thank Center for Gardening and Horticulture, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences for providing the plant material.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession number OK649927. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA816672, SRR18335031, and SAMN26688270, respectively.
Additional information
Funding
References
- Abdel-Hameed UK. 2014. Morphological phylogenetics of Bignoniaceae Juss. Beni-Suef Univ J Basic Appl Sci. 3:172–177.
- Chen Q, Wu XB, Zhang DQ. 2020. Comparison of the abilities of universal, super, and specific DNA barcodes to discriminate among the original species of Fritillariae cirrhosae bulbus and its adulterants. PLos One. 15(2):e0229181.
- Corlett RT. 2004. Flower visitors and pollination in the Oriental (Indomalayan) Region. Biol Rev Camb Philos Soc. 79(3):497–532.
- Darwin C. 1876. The effects of cross and self fertilisation in the vegetable kingdom. London (UK): John Murray.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kubitzki K, editor. 2004. The families and genera of vascular plants VII. Flowering plants dicotyledons: Lamiales (except Acanthaceae including Avicenniaceae). Berlin (Germany): Springer Science & Business Media.
- Milet-Pinheiro P, Carvalho AT, Kevan PG, Schlindwein C. 2009. Permanent stigma closure in Bignoniaceae: mechanism and implications for fruit set in self-incompatible species. Flora Morphol Distribution Funct Ecol Plants. 204(1):82–88.
- Olmstead RG, Zjhra ML, Lohmann LG, Grose SO, Eckert AJ. 2009. A molecular phylogeny and classification of Bignoniaceae. Am J Bot. 96(9):1731–1743.
- Shi LC, Chen HM, Jiang M, Wang LQ, Wu X, Huang LF, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Spangler RE, Olmstead RG. 1999. Phylogenetic analysis of bignoniaceae based on the cpDNA gene sequences rbcL and ndhF. Ann Mo Bot Gard. 86(1):33–46.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1312.
- van Steenis CGGJ. 1977. Bignoniaceae. In: van Steenis CGGJ, editor. Flora Malesiana. Series 1, Vol. 8. Alphen Aan Den Rijn (Netherlands): Sijthoff & Noordhoff International Publishers; p. 114–186.
