Abstract
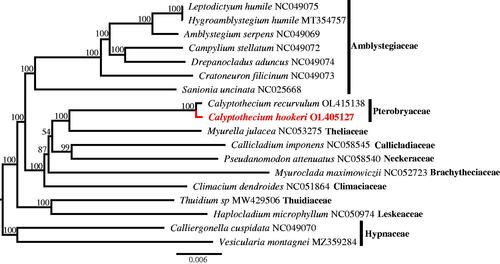
In this study, the complete chloroplast chloroplast genome of Calyptothecium hookeri was studied and reported. The size of the entire chloroplast genome was 124,401bp in length, comprising of two inverted repeats (IRa and IRb, 9371 bp respectively) separated by one large single copy (LSC: 87,126 bp) and one small single copy (SSC: 18,533 bp). The GC content of the genome sequence was 41.28%. A total of 126 functional genes were predicted, consisting of 82 protein-coding genes, 36 tRNA genes, and eight rRNA genes. Phylogenetic analysis showed that the two Pterobryaceae species C. hookeri and C. recurvulum clustered in one clade, which is sister to the Theliaceae species Myurella julacea.
The genus Calyptothecium was established by Mitten (1868) and currently comprises ca. 30 species (Frey and Stech Citation2009), which are widely distributed in the pantropical and Southeast Asia. It is characterized by large plant with creeping stem, cordate to auricular leaf base and oblong-ovate leaf with single costa. Calyptothecium hookeri (Mitt.) Broth. was described as Meteorium hookeri based on four specimens collected from Khasia and Sikkim in India (Mitten Citation1859). It was transferred to Calyptothecium by Brotherus in Citation1906. In this study, we report the first complete plastome of C. hookeri based on Illumina paired-end sequencing data, for further studies on phylogeny of the Hypnales.
The fresh leaves of C. hookeri were collected from Kashang village (22°59′29.4″N, 104°3′23.18″E), Maguan County, Yunnan province, China. The voucher specimen (Wei Han HW802) was deposited in National Herbarium (PE, Ningning Yu, [email protected]), Institute of Botany, the Chinese Academy of Sciences (IBCAS) under the barcode number PE01760194. DNA libraries were prepared with an insert size of 350 bp using NEBNext Ultra DNA Library Prep Kit (NEB, USA). Paired-end sequencing (150 bp reads) was performed on an Illumina NovaSeq 6000 platform (San Diego, CA) at Novogene Co. Ltd. (Beijing, China). In total, 7.03 Gb raw sequence data were obtained with a number of raw reads 23,429,275. The complete chloroplast genome was assembled using the software NOVOWrap (Wu et al. Citation2021). The genes were annotated using the GeSeq web application (Tillich et al. Citation2017) and modified manually. The scientific research administrative of IBCAS authorized this study, which included sample collection, and it complied with national and local laws.
The plastome of C. hookeri is 124,401bp in length, containing two inverted repeats (IRa and IRb, 9,371 bp respectively) separated by one large single copy (LSC: 87,126 bp) and one small single copy (SSC: 18,533 bp). The total GC content was 41.28% and 126 functional genes were predicted, including 82 protein-coding genes, 36 tRNA genes and eight rRNA genes.
The complete chloroplast genome of C. hookeri, together with 17 species in the Hypnales, were used for phylogenetic analysis. The sequence alignment was performed using MAFFT v7.307 (Katoh and Standley Citation2013). Phylogenetic relationships of C. hookeri were estimated via IQ-TREE web server (http://iqtree.cibiv.univie.ac.at/) (Trifinopoulos et al. Citation2016). The phylogenetic tree showed that C. hookeri and C. recurvulum clustered in one clade, which is sister to Myurella julacea ().
Authors’ contributions
Conception and design: Ningning Yu and Yu Jia; Analysis and interpretation: Wei Han; Drafting: Wei Han and Ningning Yu; Revising: Ningning Yu and Yu Jia; All authors reviewed the manuscript.
Acknowledgments
The authors acknowledge Dr. Ping Wu from Institute of Botany, the Chinese Academy of Sciences.
Disclosure statement
The authors alone are responsible for the content and writing of the paper. The authors report no conflicts of interest.
Data availability statement
The genome sequence data that obtained at this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov) under accession no. OL405127. The associated BioProject, SRA, and BioSample numbers are PRJNA775849, SRR17271037 and SAMN23176158, respectively.
Additional information
Funding
References
- Brotherus VF. 1906. Nat. Pflanzenfam. Leipzig: Engelmann; Vol. I(3), p. 839.
- Frey W, Stech M. 2009. Bryophyta (Musci, Mosses). In: Frey W, Stech M, Fischer E, editors. Syllabus of plant families: bryophytes and seedless vascular plants. Stuttgart (Germany): Borntraeger.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Mitten W. 1859. Musci Indiae Orientalis, an enumeration of the mosses of the East Indies. J Proc Linn Soc Bot Suppl. 1:1–171.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.
- Wu P, Xu C, Chen H, Yang J, Zhang XC, Zhou SL. 2021. NOVOWrap: an automated solution for plastid genome assembly and structure standardization. Mol Ecol Resour. 21(6):2177–2186.