Abstract
Coreoperca liui as an approximate species of the genus Siniperca, provides an important source for the genetic diversity of the mandarin fish, which is valuable for the protection of biodiversity and utilization of germplasm resources. The complete mitochondrial genome of C. liui is 16,482 bp long and it consists of 13 protein-coding genes (PCGs), two ribosomal RNA genes, 22 transfer RNA genes, and a control region (D-loop). Phylogenetic analysis using the maximum-likelihood method, based on 13 PCGs and two rRNA from 13 species produced three major clades. The phylogenetic tree showed that C. liui is most closely related to Coreoperca whiteheadi. Our results provide useful information for understanding the phylogeny of the genus Coreoperca, as well as for conducing conservation studies of Sinipercidae and related species.
Coreoperca liui (Cao and Liang Citation2013) is a member of the order Perciformes and it belongs to the family Sinipercidae, one of the most diverse groups of mandarin fish. They usually live in streams with rapid water flow and good water quality (Cao and Liang Citation2013). The distribution of C. liui is in the southeast coastal areas of China, where the species is mainly distributed in the lower reaches of the Yangtze River, Pearl River, and other river basins (Li Citation1991; Song et al. Citation2017). Its habitat is rapidly deteriorating owing to industrial wastewater discharge and hydropower dam construction (Lin et al. Citation2019; Liu et al. Citation2019). There were only few reports on the genetic research of C. liui, and no report on its complete mitogenome. The analysis of mitochondrial DNA fragment and complete mitogenome have been successfully applied in fish identification, phylogenetic analysis, and population biology (Billington and Hebert Citation1991).
In this study, the complete mitogenome of C. liui was determined through Illumina Hiseq sequencing (GenBank accession number: MZ964309). The fish was sampled from Qiandaohu Lake, located in Zhejiang Province, China (29.37°N, 118.73°E). The sample was preserved in 95% ethanol and deposited at Huzhou University (www.zjhu.edu.cn, Yixiang Zhang, [email protected]) under the voucher number HZ202010211. The total genomic DNA was extracted from fish muscles following the method described in Tang et al. (Citation2008), and then sequenced using Illumina HiSeq4000 (Han et al. Citation2020). After sequencing, the complete mitogenome was assembled through NOVOPlasty (https://github.com/ndierckx/NOVOPlasty), and annotated using MITOS (http://mitos2.bioinf.uni-leipzig.de/index.py) (Dierckxsens et al. Citation2016; Donath et al. Citation2019). It was also annotated using Coreoperca whiteheadi (KJ149811.1 in GenBank) as a reference (Lv et al. Citation2016).
The entire mitochondrial genome of C. liui is a circular molecule with a length of 16,482 bp, which consists of 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, 22 transport RNA (tRNA) genes, and a control region (D-loop). The overall nucleotide composition is 28% A, 26.69% T, 29.24% C, and 16.07% G. The content of A + T is 54.69%, which shows an obvious AT preference, and the gene content and arrangement are similar to the mitochondrial genome of typical vertebrates. ND6 and eight tRNAs (tRNAGln, RNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, tRNAPro) are encoded on the L-strand, while the others are encoded on the H-strand. Two types of start codons (ATG, GTG) and four types of stop codons (TAG, TAA, TA–, T––) were used in the 13 PCGs. Most of the PCGs start with ATG, while the codon of COI is GTG. Six PCGs were terminated with the complete stop codons TAA or TAG; ATPase6 and COIII were terminated with incomplete codon (TA–); and five protein coding genes (ND2, ND3, ND4, Cytb, COII) were terminated with the incomplete codon (T––), which was similar to past reports on the mitochondrial genes of other fishes. The truncated stop codons TA– and T–– are very common in animals, which are presumably completed as TAA by post-transcriptional polyadenylation (Boore Citation1999).
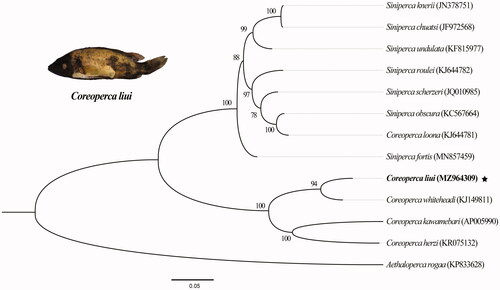
Comparisons between C. liui and 12 other species using the COI gene and 13 PCGs from NCBI (https://www.ncbi.nlm.nih.gov/nuccore/MZ964309/) showed that the sequence identity between C. liui and C. whiteheadi is the highest at approximately 87.96%, and the sequence identities with C. herzi, C. kawamebari, and Siniperca scherzeri are 87.69%, 86.07%, and 83.46%, respectively (Yamanoue et al. Citation2007; Chu et al. Citation2013; Park et al. Citation2016). The molecular phylogenetic tree was constructed based on two rRNA and 13 protein coding genes from C. liui and 12 others related species of the subfamily Serranidae with the species Aethaloperca rogaa (Forsskål, 1775) as an outgroup, using the maximum-likelihood method with 1000 replicates in IQ-tree 2.1.2 (http://www.iqtree.org/). The most suitable nucleotide substitution pattern (TPM2 + F + R3) was selected on the basis of the BIC (Minh et al. Citation2020) (). According to our results, C. liui had a closer relationship with C. whiteheadi than with the other four species of Coreoperca, in agreement with the result of the COI-based BLAST analysis in NCBI. This mitochondrial genome provides important genomic information on the genus Coreoperca that may contribute to biodiversity protection and phylogenetic analysis of Serranidae.
Ethics statement
All methodologies used in the experiments of this study were approved by the Committee of Ethics and Animal Welfare of Huzhou University and complied with local wildlife protection laws.
Author contributions
Fangyuan Guan conceived the research and wrote and edited the manuscript. Qiang Sheng conceived the research and performed the experiments. Yixiang Zhang and He Lv analyzed the data. Yingying Wang conceived and designed the research. All authors contributed to the article and approved the version to be published. All authors agreed to be accountable for all aspects of the work.
Acknowledgements
We are grateful to Zhi Jin and Tianjiang Chu for assistance in sample collection. We thank Natalie Kim, PhD, from Liwen Bianji (Edanz) (www.liwenbianji.cn/), for editing the English text of a draft of this manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, under the accession no. MZ964309. The associated BioProject, Bio-Sample, and SRA accession numbers are PRJNA825346, SAMN27512414, and SRR18709848, respectively.
Additional information
Funding
References
- Billington N, Hebert PD. 1991. Mitochondrial DNA diversity in fishes and its implications for introductions. Can J Fish Aquat Sci. 48(S1):80–94.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Cao L, Liang XF. 2013. A new freshwater perch species of the genus Coreoperca herzenstein (Perciformes, Serranidae, Sinipercinae) from Zhejiang Provence, China. Acta Zootaxon Sin. 38(4):891–894.
- Chu W, Chen D, Wang K, Li Y, Du S, Zhang J. 2013. Analysis of the variable sites and phylogenetic studies of complete mitochondrial DNA based on the Siniperca scherzeri (Perciformes: Sinipercidae) from four different areas. Mitochondrial DNA. 24(3):288–289.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552.
- Han YQ, Huang L, Zhu QH, Wu WJ, He AY, Shi J, Li YS. 2020. Characteristics of mitochondrial genomes of Semilabeo obscures analyzed using Illumina Hiseq 4000 high-throughput sequencing technique. Fujian J Agric Sci. 35(2):130–139.
- Li SZ. 1991. Geographical distribution of the Sinipercine fishes. Chin J Zool. 26:40–44.
- Lin PC, Wang CL, Liu F, Liu M, Liu HZ, Wang XM, Yu J, Zhu X. 2019. Current status and conservation planning of fish biodiversity in the upper Yangtze River basin in the context of hydropower development. Acta Hydrobiol Sin. 43(S1):130–143.
- Liu F, Lin PC, Liu MZ, Gao X, Wang CL, Liu HZ. 2019. Situations and conservation strategies of fish resources in the Yangtze River basin. Acta Hydrobiol Sin. 43(S1):144–156.
- Lv LY, Tian CX, Liang XF, Yuan YC, Zhao C, Song Y. 2016. The complete mitochondrial genome sequence of Coreoperca whiteheadi (Perciformes: Serranidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):301–303.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler H, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):268–274.
- Park CE, Park GS, Kwak YY, Hong SJ, Khan AB, Jung BK, Park YJ, Kim MC, Kim KH, Park HC, et al. 2016. Complete mitochondrial genome of the endemic species Korean aucha perch Coreoperca herzi (Teleostei, Centrarchiformes, Sinipercidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3493–3495.
- Song SL, Zhao J, Li C. 2017. Species delimitation and phylogenetic reconstruction of the sinipercids (Perciformes: Sinipercidae) based on target enrichment of thousands of nuclear coding sequences. Mol Phylogenet Evol. 111:44–55.
- Tang QY, Freyhof J, Xiong BX, Liu HZ. 2008. Multiple invasions of Europe by East Asian cobitid loaches (Teleostei: Cobitidae). Hydrobiologia. 605(1):17–28.
- Yamanoue Y, Miya M, Matsuura K, Yagishita N, Mabuchi K, Sakai H, Katoh M, Nishida M. 2007. Phylogenetic position of tetraodontiform fishes within the higher teleosts: Bayesian inferences based on 44 whole mitochondrial genome sequences. Mol Phylogenet Evol. 45(1):89–101.