Abstract
Lilium primulinum var. burmanicum (W. W. Smith) Stearn 1948 is an ornamental and medicinal plant that has an extremely limited distribution in Yunnan, China. Here, we obtained the complete plastome of L. primulinum var. burmanicum via next-generation sequencing, and conducted phylogenomic analyses with existing species from Lilium. The total length of L. primulinum var. burmanicum was 152,206 bp with a typical quadripartite structure. The whole plastome contained a pair of inverted repeats (IRa/IRb; 26,399 bp) which divided a large single-copy (LSC; 81,854 bp) and a small single-copy (SSC; 17,563 bp). The average GC content among the whole plastome sequence and the LSC, SSC, and IR regions were 37%, 34.8%, 30.6%, and 42.5%, respectively. There were 134 genes detected from the whole plastome sequence, including 87 protein-coding genes, 39 tRNAs, and 8 rRNAs. Phylogenetic analyses using maximum likelihood showed congruent results that L. primulinum var. burmanicum together with L. primulinum var. ochraceum formed a single branch. These results demonstrate a close relationship between these variation species. The newly characterized chloroplast genome presented here will provide essential data for further phylogenomic analyses of the intraspecific relationship among Lilium species and for conservation genetics research of L. primulinum var. burmanicum.
Lilium (Liliaceae) species are cultivated as potted plants or cut flowers for their bright-colored flowers and extraordinary fragrance (Shahin et al. Citation2011). More than this, Bulbus Lilii is an important traditional Chinese drug due to its medicinal properties, such as nourishing the lung and alleviating mental stress (Chinese Pharmacopoeia Commission Citation2020). Therefore, many species of Lilium are at risk of extinction in the field due to overutilization.
Lilium primulinum var. burmanicum (W. W. Smith) Stearn, 1948, is a perennial herb that inhabits the edge of mountainous forests at elevations ranging from ∼ 1200 to 2700 m in China (Chen et al. Citation2000). According to herbarium, L. primulinum var. burmanicum had been widely distributed, with at least 15 specimen occurrence records in Yunnan. However, based on our field work during 2011–2020, only 4 populations with approximately 20 individuals per population were found. We inferred that with illegal removal from natural habitats for horticultural and medicinal use, L. primulinum var. burmanicum has likely become vulnerable and decreased in the number of populations, which may lead to extinction in the wild and should draw attention to this nearly endangered species. Here, we first reported the complete chloroplast genome of L. primulinum var. burmanicum and determined the phylogenetic analyses within Lilium species.
Samples of L. primulinum var. burmanicum were collected from Malipo County in Yunnan, China (23°09′49″N, 104°49′35.4″E), and the specimens were deposited in the Herbarium of Kunming University (YGS1107023; Genshen Yin, [email protected]). The total genomic DNA of L. primulinum var. burmanicum was extracted from dry leaves stored in silica using a Plant Genomic DNA Kit (DP305; Tiangen, Beijing) according to the manufacturer’s instructions. The quality and concentration of total DNA were determined by Nanodrop 2000 Spectrophotometers (Thermo Fisher Scientific Inc., Waltham, MA). The total genome was sequenced with paired-end reads of 150 bp on the Illumina HiSeq platform (Illumina Inc., San Deigo, CA). All raw reads were assembled using GetOrganelle software (Jin et al. Citation2020) with 47 published plastid genome sequences of the genus Lilium, referred to as “embplant_pt.” Genome annotation was performed by PGA (Qu et al. Citation2019) and GeSeq (Tillich et al. Citation2017) with the plastid genome of L. pardanthinum (GenBank accession: MG704135) as the reference. Transfer RNA (tRNA) genes were identified with the tRNAscan-SE program with default parameters (Schattner et al. Citation2005). OrganellarGenomeDRAW (OGdraw) was used to draw a circular chloroplast genome map (Marc et al. Citation2018). Finally, the whole chloroplast genome of L. primulinum var. burmanicum was deposited into GenBank (Accession no. MZ188968).
The complete chloroplast genome of L. primulinum var. burmanicum showed a typical quadripartite cycle with a length of 152,309 bp, containing two inverted repeat (IR) regions of 26,394 bp each, which were separated by a large single-copy (LSC, 81,956 bp) region and a small single-copy (SSC, 17,565 bp) region, similar to L. primulinum var. ochraceum (accession no.: KY748298). The GC content of the complete plastid genome was 37%, whereas the corresponding values of the LSC, SSC, and IR regions were 34.8%, 30.6%, and 42.5%, respectively. We detected 134 genes from the cp genome sequence, consisting of 87 protein-coding genes, 39 tRNA genes, and 8 rRNA genes. Among these, seven protein-coding genes, eight tRNA genes, and four rRNA genes were duplicated. However, the protein-coding genes cemA and ndhD were annotated in duplicate in a previous study carried out by Du et al. (Citation2017). In both L. primulinum species, 25 introns contained genes.
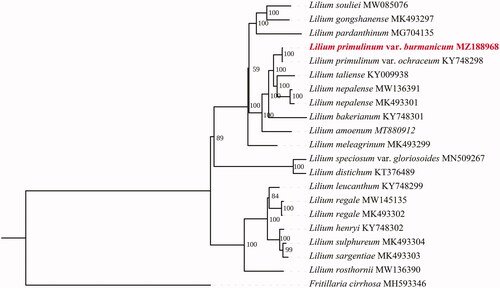
Maximum-likelihood (ML) phylogeny was inferred by IQ-tree software (Minh et al. Citation2020) from the total chloroplast genome sequence of 19 species, including a representative of Fritillaria that was used as outgroup (). The best-fit models, GTR + F + I + G4, were used in the analysis with 1000 bootstrap replicates. The phylogenetic results showed a close relationship of L. primulinum var. burmanicum with L. primulinum var. ochraceum. There was only one singleton variable site (G/C) between these two species, located in the intergenic regions of trnT-UGU and trnL-UAA. In addition to the variable site, five InDel events were also detected among the single-nucleotide microsatelliteregions. Information on the complete chloroplast genome of L. primulinum var. burmanicum could provide valuable insight into conservation and exploitation efforts for this endangered species.
Ethics of experimentation statement
The authors ensure that the plant material (Lilium primulinum var. burmanicum) reported in submitted papers was handled in an ethical and responsible manner and the study was conducted in full compliance with all relevant codes of experimentation and legislation. The total genomic DNA extraction from dry leaves saved in silica does not harm the growth of plants. In addition, the plant samples used in this research were not collected from private land or a nature reserve, so ethics approval was not required for this research.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank under accession no. MZ188968 (https://www.ncbi.nlm.nih.gov/nuccore/MZ188968.1/). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA729634, SRR14520563, and SAMN19136515, respectively.
Additional information
Funding
References
- Chen XQ, Liang SY, Xu JM, Tamura MN. 2000. Flora of China (vol. 24). Beijing/St. Louis (MO): Science Press/Missouri Botanical Garden Press.
- Chinese Pharmacopoeia Commission. 2020. Pharmacopoeia of the People's Republic of China. Beijing: China Medical Science Press.
- Du YP, Bi Y, Yang FP, Zhang MF, Chen XQ, Xue J, Zhang XH. 2017. Complete chloroplast genome sequences of Lilium: insights into evolutionary dynamics and phylogenetic analyses. Sci Rep. 7(1):5751.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Marc L, Oliver D, Sabine K, Ralph B. 2018. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(W1):W575.
- Minh BQ, Schmidt H, Chernomor O, Schrempf D, Woodhams M, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server):W686–689.
- Shahin A, Arens P, Van Heusden AW, Van Der Linden G, Van Kaauwen M, Khan N, Schouten HJ, Van De Weg WE, Visser RGF, Van Tuyl JM. 2011. Genetic mapping in Lilium: mapping of major genes and quantitative trait loci for several ornamental traits and disease resistances. Plant Breeding. 130(3):372–382.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.