Abstract
Briggsia chienii W. Y. Chun 1946 is an endemic herbaceous perennial species distributed in southern China. In this study, we firstly characterized the complete chloroplast genome sequence of B. chienii and provided new molecular resources for promoting its conservation and taxonomic assignment. Its complete chloroplast genome is 154,082 bp in length and contains the typical quadripartite structure of angiosperm plastome, including two inverted repeat (IR) regions of 25,447 bp, a large single-copy (LSC) region of 85,035 bp, and a small single-copy (SSC) region of 18,153 bp. The plastome contains 114 genes, consisting of 80 protein-coding genes, 30 tRNA gene, and 4 rRNA genes. The overall GC content in the plastome of B. chienii is 37.4%, which is lower than lots of angiosperm plastome. The phylogenetic result indicated that B. chienii exhibited the closest relationship with Oreocharis cotinifolia W. T. Wang 1983, and provided new information for the phylogeny relationship of genus Briggsia.
Briggsia chienii W. Y. Chun 1946 is an herbaceous perennial plant belonging to family Gesneriaceae. It is a Chinese endemic plant, and only can be found on moist rocks and grasses between 500 and 1000 meters of southwestern Zhejiang, southern Anhui, and eastern Jiangxi. The plants of B. chienii is stemless, all leaves are basal and stalked, and corolla is purple-red or purple with inside purple spots. B chienii is a medicinal plant that can also be utilized as a garden or indoor potted plant. The genus Briggsia was established in 1919 by Craib (Craib Citation1919), and have been considered to consist of 22 species and three varieties in the world, one of which is distributed in Sikkim, and the other three are shared by China, Myanmar, Bhutan, India, and Vietnam, and the remaining 18 species are endemic to China (Wang et al. Citation1998). According to previous studies, this genus should not be recognized as a valid group, and all of its species should be moved to other genera, including Glabrella, Loxostigma, and Oreocharis. The synonym of B. chienii should be Oreocharis chienii (Möller et al. Citation2014). However, there are limited researches on the genus Briggsia, and still insufficient morphological and molecular evidences to transfer all its member species to other gnus, especially for species B. chienii. Here, we reported and characterized the first complete chloroplast genome of B. chienii based on high throughput sequencing technology, and reconstructed the phylogeny relationships by utilizing the published chloroplast genome sequences of Gesneriaceae.
The original plants of B. chienii were collected from Fuxi village, Tangkou Town, Huangshan City in Anhui Province (N30°07′, E118°12′). And the plants were collected and identified by Xinlei Zhao ([email protected], this sample was neither collected from a protected area nor listed on any endangered species list, such as CITES. And, under current law, what is not prohibited is what is allowed to be collected). These materials are deposited in the Institute of Medicinal Plant Development herbarium (herbarium code ‘IMD,’ NYBG: https://www.nybg.org/, Linchun Shi: [email protected]) with the voucher number as HPAA0003. Genomic DNA was extracted from the leaf tissues using the plant genomic DNA extraction kit [Tiangen Biochemical Technology (Beijing) Co., Ltd, China] according to the protocol provided by the manufacturer with some modifications (Zhang et al. Citation2021). The quantity and quality of the total DNA were examined using Qubit 4.0 (Thermo Fisher Scientific Inc., USA). The sequencing library was constructed according to the TruSeq DNA PCR-free library preparation guide. The Illumina NovaSeq platform was employed to conduct the high-throughput sequencing, and approximately 1.8 Gb of clean data was generated with 150 bp paired-end read lengths. Trimmonmatic v0.38 was employed to remove the adapters and low-quality reads with the parameters ‘TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36’ (Bolger et al. Citation2014). The remaining reads were assembled into the complete chloroplast genome by using the organelle assembler GetOrganelle v1.7.3.5 with the parameters ‘-R 15 -k 21,45,65,85,105 -F embplant_pt’ (Jin et al. Citation2020), and validated by reads mapping using bowtie2 (Langmead and Salzberg Citation2012). The protein-coding, rRNA, and tRNA genes of B. chienii chloroplast genome were annotated by using the CPGAVAS2 online webserver (www.herbalgenomics.org/cpgavas2) (Shi et al. Citation2019). The complete chloroplast genome sequence of B. chienii was submitted to GenBank (Accession number: MZ868555).
The length of the B. chienii complete chloroplast genome sequence was 154,082 bp, and the total GC content was 37.4%. The genome displayed a typical quadripartite structure, including large single-copy (LSC) region (85,035 bp), small single-copy (SSC) region (18,153 bp) and a pair of inverted repeats IR regions (25,447 bp). There are 114 unique genes in the whole chloroplast genome sequence of B. chienii, including 80 protein-coding genes, 30 represented tRNA genes, and four denoted rRNA genes (rrn23S, rrn16S, rrn5S, and rrn4.5S). Among them, 19 genes were annotated as containing introns. Nine protein-coding genes and seven tRNA genes (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnR-UCU, trnV-UAC) contained one intron, and three protein-coding genes (clpP, ycf3, and rps12) contained two introns. In addition, small exons were annotated in petB, petD, and rpl16 genes, and the length of their small exons were 6 bp, 8 bp and 9 bp, respectively. Moreover, rps12 was identified as a trans-splicing gene.
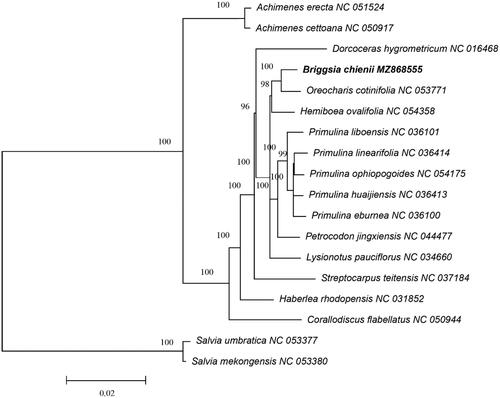
To confirm the phylogenetic position of B. chienii in Gesneriaceae, a total of 18 complete chloroplast genomes were used for phylogenetic analysis based on the Maximum Likelihood (ML) method using RAxML v8.2.12 with 1000 bootstrap replicates (Stamatakis Citation2014). Salvia mekongensis E. Peter 1936 and S. umbratica Hance 1884 were used as the out groups. The phylogenetic result indicated that B. chienii exhibited the closest relationship with Oreocharis cotinifolia (), and was consistent with the prior taxonomic suggestion (Möller et al. Citation2014). This study provided a useful molecular resource for its conservation and the phylogenetic studies of genus Briggsia.
Author contributions
X.X., J.L. and X.D. conceived and designed the experiments; M.S and L.S. performed the experiments; M.S and C.S. analyzed the data; M.S., L.S. and W.Z. contributed reagents/materials/analysis tools; X.X., J.L. and X.D. wrote the paper.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genom esequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ868555. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA682118, SRR16930608, and SAMN23005362, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Craib WG. 1919. Gesneracearum novitates, vol 11. Edinburgh: Notes Roy Bot Gard Edinburgh: pp. 233–254.
- Jin J-J, Yu W-B, Yang J-B, Song Y, de Pamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359.
- Möller M, Chen WH, Shui YM, Atkins H, Middleton DJ. 2014. A new genus of Gesneriaceae in China and the transfer of Briggsia species to other genera. Gardens’ Bulletin (Singapore). 66(2):195–205.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang W, Pank LZ, Weitzman AL, Skog LE. 1998. Gesneriaceae. Flora of China. St. Louis: Science Press, Beijing & Missouri Botanical Garden Press.
- Zhang G, Wang H, Jiang J, Wang Q, Li B, Shi L, Wei J. 2021. The complete chloroplast genome of Bupleurum marginatum var. stenophyllum (H. Wolff) Shan & Yin Li (Apiaceae), a new substitution for Chinese medicinal material, Bupleuri Radix (Chai hu). Mitochondrial DNA B Resour. 6(2):441–443.