Abstract
The complete chloroplast genome of Pulsatilla campanella Fischer ex Krylov was sequenced and reported for the first time. The length of the entire circular genome was 162,322 bp, and the GC content was 37.4%. There were 133 genes annotated, including 89 known protein-coding genes, 36 tRNAs, and 8 rRNAs. The complete chloroplast genome of P.campanella has consisted of two inverted repeat regions (IRs), a large single-copy region (LSC 82,087 bp), and a small single-copy region (SSC 17,497 bp). The phylogenetic tree was built based on 29 species, using the maximum-likelihood method. The results showed that P.campanella was clustered on the same branch with a variety of Pulsatilla plants. The data reveal the genetic relationship between the selected species and provide information for subsequent plant classification.
Pulsatilla campanella Fischer ex Krylov (FI. Ajan. 30. 1859) is an early spring plant with purple perianths, and its flower nodding before and at anthesis resembles a bell. It is mainly distributed across western Xinjiang, China, in addition to northern Mongolia and Russia's Central Asia. P. campanella is an important species in the genus Pulsatilla. Many species of the genus Pulsatilla have been used as a traditional medicine named ‘Bai Tou Weng,’ which has a long history and significant therapeutic effect in clinical practice (Kumar et al. Citation2008). The roots of P. campanella are used to treat bacillary dysentery and lymphatic tuberculosis in local Chinese herbal medicine records (Jiangsu New Medical College Citation1977; Li et al. Citation2019). Triterpene saponins proved to be the main active substances in P. campanella (Li et al. Citation1990). At present, there are many studies on chemical composition and pharmacological activities (Xu et al. Citation2012; Zhang et al. Citation2019), and the chloroplast DNA fragments for species identification have also been reported (Li et al. Citation2019). However, it is the first time to report on the sequencing of the complete chloroplast genome of P. campanella.
The experimental materials were collected from Tekes, Ili Kazak Autonomous Prefecture, Xinjiang, China (E 81°89′30″, N 43°26′98″), and were identified as P. campanella by professor Tingguo Kang (Liaoning University of Traditional Chinese Medicine, Dalian, China). We used 200 mg of fresh leaves to extract total genomic DNA using the modified CTAB method (Doyle and Doyle Citation1987). The total genomic DNA was constructed in a sequencing library with a 350 bp insert using the NexteraXT DNA library preparation kit (Illumina, San Diego, CA, USA), and double-terminal sequencing was performed on the library using the Illumina Novaseq 6000 sequencing platform. The raw data was edited using NGS QC Tool Kit v2.3.3 (Patel and Jain Citation2012). High-quality reads were assembled into chloroplast genome using a de novo assembler SPAdes v3.11.0. (Bankevich et al. Citation2012). Finally, it was annotated by PGA (Qu et al. Citation2019) with Pulsatilla dahurica (MK860685) as reference genome. All experimental contents were approved by the School of Pharmacy, Liaoning University of Traditional Chinese Medicine, Dalian, China (20200112). All operations were under guidelines from the Specification on Good Agriculture and Collection Practices for Medicinal Plants (GACP; Number: T/CCCMHPIE 2.1-2018). The voucher specimen and genomic DNA were kept in the herbarium of Liaoning University of Traditional Chinese Medicine (Xinlei Zhao and Liang Xu, [email protected], P.campanella number: 10162200525972LY). The sequencing data were assembled using ABySS v2.0.2 (http://www.bcgsc.ca/platform/bioinfo/software/abyss). The protein-coding sequences of chloroplast were compared with the known protein databases (eggNOG, GO, KEGG, NR, Swiss-Prot) to predict protein-coding genes.
The entire genome is 162,322 bp in length. It has a typical circular structure, with a GC content of 37.4%. The genome contains 133 genes, including 89 known protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The total length of the coding gene is 92704 bp, accounting for 57.11% of that total length of the genome. Besides, 17 genes (rnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, TrnA-UGC, ndhA, clpP, ycf3) containing 19 introns has been found.
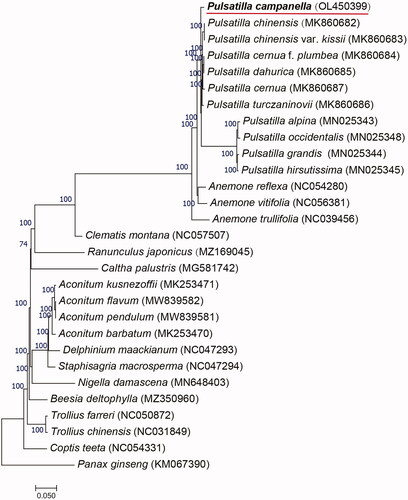
The phylogenetic tree is a commonly used method to visualize the evolutionary relationship between species. The complete chloroplast genomes of 29 species (including P. campanella) were selected to build a phylogenetic tree, using the maximum likelihood method and the model TVM + F+R4 in IQ-TREE 1.6.12 (Nguyen et al. Citation2015) with 1000 bootstrap replicates(). In the process, Panax Ginseng existed as an outgroup. The phylogenetic tree showed that Panax Ginseng was distant from other species, and P. campanella was clustered on the same branch with a variety of pulsatilla plants, which confirms the authenticity and reliability of the results. The data will not only provide a basis for analyzing the phylogenetic position of P. campanella, but also lay a theoretical foundation for the systematic classification of Ranunculaceae.
Author contributions
Hefei Xue: Analysis of data, conception and drafting for the work. Yueyue Song: Revising critically for important intellectual content. Yanyun Yang and Che Bian: Acquisition and analysis of data. Liang Xu and Tingguo Kang: Final approval of the version to be published.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession NO. OL450399. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA778819, SRX13107158 (Illumina), and SAMN22999511, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Jiangsu New Medical College. 1977. The dictionary of traditional Chinese medicines; p. 704–705. Shanghai: Shanghai Science and Technology Publishers.
- Kumar S, Madaan R, Farooq A, Sharma A. 2008. The genus Pulsatilla: a review. Pharmacogn Rev. 2(3):116–123.
- Li QJ, Wang X, Wang JR, Su N, Zhang L, Ma YP, Chang ZY, Zhao L, Potter D. 2019. Efficient identification of Pulsatilla (Ranunculaceae) using DNA barcodes and micro-morphological characters. Front Plant Sci. 10:1196.
- Li XC, Wang DZ, Wu SG, YANG CR. 1990. Triterpenoid saponins from Pulsatilla campanella. Phytochemistry. 29(2):595–599.
- Nguyen LT, Schmidt HA, Haeseler AV, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Xu QM, Shu Z, He WJ, Chen LY, Yang SL, Yang G, Liu YL, Li XR. 2012. Antitumor activity of Pulsatilla chinensis (Bunge) Regel saponins in human liver tumor 7402 cells in vitro and in vivo. Phytomedicine. 19(3–4):293–300.
- Zhang WD, Jiang HH, Yang JX, Song GS, Wen D, Liu WQ, Jin MM, Wang Q, Du YF, Sun Q, et al. 2019. A high-throughput metabolomics approach for the comprehensive differentiation of four Pulsatilla Adans herbs combined with a nontargeted bidirectional screen for rapid identification of triterpenoid saponins. Anal Bioanal Chem. 411(10):2071–2088.