Abstract
Besides being a common weed, the presence of Echium plantagineum L. in food and feed commodities can represent a safety hazard due to their content in pyrrolizidine alkaloids. In this study, the complete chloroplast of E. plantagineum isolate BPTPS251 is described, being the first available plastome from an isolate belonging to the Echium genus. The chloroplast genome is 149,776 bp in length with 37.5% GC content, displaying a quadripartite structure that contains a pair of inverted repeats regions (25,754 bp each), separated by a large single-copy (80,978 bp) and a small single-copy (17,290 bp) regions. A total of 131 genes were predicted, including 37 tRNA genes, 8 rRNA genes, and 86 protein-coding genes. The phylogenetic analysis confirmed the placement of E. plantagineum under the Boraginaceae family, belonging to the Boraginales order. This study will contribute to conservation, phylogenetic, and evolutionary studies, as well as DNA barcoding applications for food and feed safety purposes.
Echium L. (viper’s buglosses) is a genus of flowering plants (angiosperms) in the Boraginaceae family, including ca. 70 species distributed in North Africa, Continental Europe, Asia Minor, and Macaronesia Islands. Some viper’s buglosses were introduced elsewhere (Australia, South Africa, and the Americas), with some garden species (e.g., Echium candicans L.fil.) being found as alien escapes. Most species are annuals (e.g., Echium vulgare L.), rosette biennials (e.g., Echium wildpretii H.Pearson ex Hook.fil. in Tenerife, the Canaries), or seldom woody shrubs (e.g., Echium vulcanorum A.Chev. in Fogo Island, Cape Verde) (WFO Citation2022).
Echium plantagineum L. (Citation1771) (Lady Campbell-weed) is a common weed in the European and Australian agricultural and waste ground areas (GBIF Secretariat Citation2021). Pharmacological activity of Echium include antimicrobial, antitumor, antiviral, and anti-inflammatory effects, mostly derived from shikonins found in the root (Wang et al. Citation2022). Seeds are also an important source of alpha-linolenic acid used in dietary supplements (Kitessa et al. Citation2011). Nectar of E. plantagineum may be important in late-winter honey production and as food for pollinators. It may also be hepatotoxic to cattle if chronically grazed due to the presence pyrrolizidine alkaloids (Moreira et al. Citation2020).
The Portuguese material of E. plantagineum analyzed, isolate BPTPS251, was collected from a wild population in Oeiras municipality in Portugal (Collection date: 20 May 2021; Location: 38.69846 N 9.31721 W) with a specimen being conserved at the LISE Herbarium (INIAV, Oeiras, Portugal; Jorge Capelo: [email protected]) under the voucher LISE: 96329 (Identified by: Jorge Capelo).
Total genomic DNA was extracted from young leaves, frozen in liquid nitrogen immediately after collection and kept at −80 °C, using an adaptation of the Doyle and Doyle (Citation1987) methodology. The obtained DNA was sheared by sonication using a Bioruptor (Diagenode), libraries were prepared with the NEBNext Ultra II DNA Library Prep Kit (New England Biolabs), and 150 bp paired-end sequencing was performed on an Illumina MiSeq platform using a v2 chemistry kit.
High-quality reads were used to assemble the complete chloroplast genome (sequence coverage: 582×) using the GetOrganelle pipeline (v1.7.5) (Jin et al. Citation2020), following the typical recipe suggested for Embryophyta plant plastome assembly (https://github.com/Kinggerm/GetOrganelle) with the additional option ‘-w 127’. The plastome annotation was performed using the GeSeq tool (Tillich et al. Citation2017) with a subsequent manual curation using Geneious Prime 2022.0.1 (https://www.geneious.com).
The chloroplast genome of E. plantagineum isolate BPTPS251 (GenBank accession number: OL335188) is 149,776 bp in length with 37.5% GC content, displaying a quadripartite structure that contains a pair of inverted repeat (IR) regions (25,754 bp, GC content 43.0%), separated by a large single-copy (LSC) region (80,978 bp, GC content 35.5%) and a small single-copy (SSC) region (17,290 bp, GC content 31.0%). A total of 131 genes were predicted, including 37 tRNA genes, 8 rRNA genes, and 86 protein-coding genes.
The phylogenetic analysis was performed using the concatenated sequences coding for the shared proteome extracted from all 11 verified and complete chloroplast genomes belonging to the Boraginales order available in GenBank (Accession date: 4 December 2021) and from the complete chloroplast genome of E. plantagineum obtained in this study. The sequences were aligned using MAFFT v7.450 (Katoh and Standley Citation2013) and further analyzed with the IQ-TREE 2 software package (Minh et al. Citation2020). The best-fit substitution model (GTR + F+R3 chosen according to the Bayesian Information Criterion) was selected according to ModelFinder (Kalyaanamoorthy et al. Citation2017), followed by a tree reconstruction () using IQ-TREE (Nguyen et al. Citation2015) using ultrafast bootstrap with UFBoot (10,000 replicates) (Hoang et al. Citation2018). The outgroup was Salvia officinalis L. (NC_038165) from the Lamiaceae family belonging to the Lamiales order.
Figure 1. Maximum-likelihood tree inferred from the sequences coding for the shared proteome from Echium plantagineum isolate BPTPS251 and all 11 verified and complete chloroplast genomes belonging to the Boraginales order available in GenBank (Accession date: 2021.12.04). Numbers attached to the branches show the SH-aLRT and the UFBoot2 percent supports (SH-aLRT/UFBoot2). Salvia officinalis (Lamiales) was used as the outgroup.
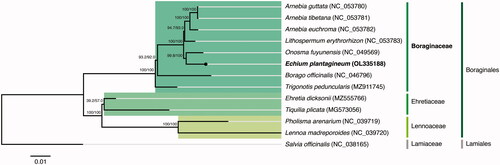
The maximum likelihood tree showed that E. plantagineum is placed under the Boraginaceae family, belonging to the Boraginales order, and has a closer relationship with Onosma fuyunensis He & Liu with 99.8%/100% support (SH-aLRT/UFBoot2). The phylogenetic analysis performed with the alignments of the Boraginales complete chloroplast genomes also supports the same tree result.
This complete chloroplast genome will contribute to conservation, phylogenetic, and evolutionary studies, as well as DNA barcoding applications for food and feed safety purposes that target the detection of pyrrolizidine alkaloid-producing species.
Ethical approval
The species described and studied in this manuscript is not under legal protection status, either by national or European Union legislation, namely the 92/43/CEE Directive. A careful nondestructive collection protocol for voucher sampling was followed to guarantee the full future reproductive viability of the studied plant population.
Author contributions
The authors had the following contribution to the paper: FBG – conception and design; JC – collection and taxonomic identification of the studied specimen; ICL and FBG – analysis and interpretation of the data; FBG – drafting of the paper; ICL, JC, MTBC, and FBG – critical revision for intellectual content; FBG – final approval of the version to be published. All authors agree to be accountable for all aspects of the work herein presented.
Disclosure statement
No potential competing or conflict of interests was reported by the authors.
Data availability statement
The data that supports this study is openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number OL335188. The associated BioProject, BioSample, and SRA numbers are PRJNA789447, SAMN22746547, and SRR16690175, respectively.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- GBIF Secretariat. 2021. GBIF backbone taxonomy. Checklist dataset accessed via https://www.gbif.org/species/2925895. on 2021-12-10.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kitessa SM, Nichols PD, Abeywardena M. 2011. Purple Viper’s Bugloss (Echium plantagineum) seed oil in human health. In Nuts and seeds in health and disease prevention. Amsterdam: Elsevier. p. 951–958.
- Linnaeus C. 1771. Mantissa plantarum altera generum editionis VI & specierum editionis II. Stockholm: Laurentii Salvii.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Moreira R, Fernandes F, Valentão P, Pereira DM, Andrade PB. 2020. Echium plantagineum L. honey: search of pyrrolizidine alkaloids and polyphenols, anti-inflammatory potential and cytotoxicity. Food Chem. 328:127169.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang W, Jin J, Xu H, Shi Y, Boersch M, Yin Y. 2022. Comparative analysis of the main medicinal substances and applications of Echium vulgare L. and Echium plantagineum L.: a review. J Ethnopharmacol. 285:114894.
- WFO. 2022. World Flora online. [accessed 2021 Dec 10]. http://www.worldfloraonline.org/
