Abstract
The Sand Martin (Riparia riparia) belongs to Hirundinidae. In this study, the complete mitochondrial genome of R. riparia was sequenced and characterized. The genome was 17,963 bases in length (GenBank accession no. OK537984) including 13 protein-coding genes, two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and two control regions. The overall base composition of R. riparia mitogenome was 30.5% for A, 31.8% for C, 14.5% for G, and 23.2% for T. Phylogenetic analysis revealed that R. riparia was genetically closest to the species of genus Tachycineta. R. riparia mitogenome could contribute to our understanding of the phylogeny and evolution of this species.
In this study, we focus on the Sand Martin (Riparia riparia Linnaeus 1758), a threatened migratory bird (BirdLife International Citation2017), which is mostly distributed in the plains in the western, northern, and northeastern parts of Eurasia (Meklenburtsev Citation1954). The Sand Martin’s natural nesting habitats are sandy banks of rivers, streams and lakes (Cramp Citation1988). Goroshko (Citation1993) also indicates that R. riparia nests near rivers with floodplain vegetation and a rich grass cover and shrub thickets. Although the behavior ecology (Zhou et al. Citation2004; Ye et al. Citation2016; Saldanha et al. Citation2019) or genetic structure research based on ND2 (An et al. Citation2019) has been carried out in recent years, the complete mitochondrial data of R. riparia were still lacking.
In this study, we sequenced the complete mitochondrial genome of R. riparia (GenBank accession no. OK537984). The muscle sample was obtained from a wild R. riparia that died of natural causes in national wetland park of the Yellow River original course, Henan Province, China (E115°26.22′, N34°66.85′). The specimen was deposited at the College of Biology and Food, Shangqiu Normal University (Huaming Zhong; [email protected]) under the voucher number SQSW002. Genomic DNA was extracted from the sample using a DNeasy Blood and Tissue kit (Qiagen, Valencia, CA). Primers were designed according to the mitochondrial genomic sequences of closely related species. The complete mitochondrial genome sequence of R. riparia was amplified and sequenced by these primers using Sanger sequencing technology.
The complete mitochondrial genome of the R. riparia was 17,963 bp in length, contains 22 transfer RNA (tRNA) genes, 13 typical protein-coding genes, two ribosomal RNA (rRNA) genes (12S rRNA and 16SrRNA), and two control region (CR) (D-loop1 and D-loop2). This gene arrangement is similar to that found in other passerines (Shuo et al. Citation2016; Huang et al. Citation2019). The base composition of mtDNA is 30.5% A, 23.2% T, 31.8% C, and 14.5% G, so the percentage of A + T (53.7%) was slightly higher than G + C (46.3%). In 13 PCGs, the shortest one was ATP8 gene (168 bp) and the longest one was the ND5 gene (1818 bp). The usage of the start codon was mainly ATG in the most of mitochondrial protein coding genes besides the COI gene employing the GTG and the ND3 gene employing the ATA; the usage of the stop one was either complete (TAA, TAG, AGA, and AGG) or incomplete (T– –). The two CRs were 1081 bp and 1310 bp long, and they had a continuous region (1041 bp) of near identical sequence (>99% sequence identity). The CR1 had no repeat units detected, while the CR2 had a microsatellite consisting of a series of complex cytosine–adenine repeats.
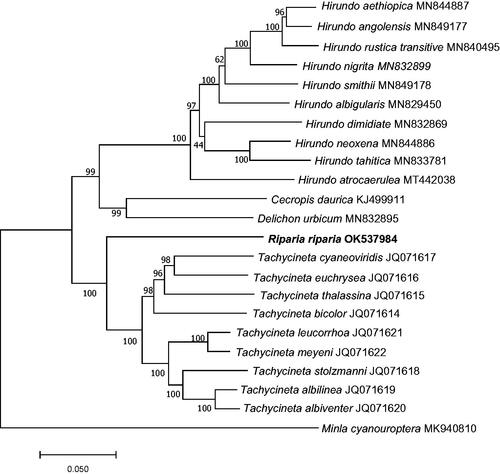
A phylogenetic tree was constructed based on 13 protein-coding gene sequences from R. riparia and 22 other Hirundinidae species using the maximum-likelihood (ML) method of MEGA version 7.0 with 1000 bootstrap replicates (Kumar et al. Citation2016). The ML tree was based on the Kimura 2-parameter model of nucleotide substitution. The results indicated that the clade of R. riparia was the sister lineage to the clade formed by the species of genus Tachycineta (). The complete mitogenomes of R. riparia will provide a better understanding of the phylogeny and evolutionary analysis of Hirundinidae.
Author contributions
The study was conceived and designed by Jianjun Peng and Chengzhong Yang; the manuscript was written by Jie Huang; the manuscript was revised by Baodong Yang and Bo Yang; the data analysis and interpretation was conducted by Jie Huang, Huaming Zhong, and Xianmeng Shi. All authors agree to be accountable for all aspects of the work.
Ethical approval
The animal study was reviewed and approved by the Ethics Committee of the Shangqiu Normal University.
Acknowledgements
We would like to acknowledge Jie Li at National Wetland Park Service of the Yellow River Original Course for collecting the sample.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OK537984. The associated BioProject and BioSample numbers are PRJNA810090 and SAMN25995379, respectively.
Additional information
Funding
References
- An BY, An Y, Wang LZ, Li B. 2019. Genetic diversity and genetic structure of Sand Martin (Riparia riparia) based on ND2. Chin J Wildlife. 40(2):377–383.
- BirdLife International. 2017. In: Staneva A, Burfield I, editors. European birds of conservation concern: populations, trends and national responsibilities; p. 1–172. Cambridge, UK: BirdLife International.
- Cramp S. 1988. Handbook of the birds of Europe, the Middle East and North Africa. The birds of the Western Palearctic. Oxford (UK): Oxford University Press.
- Goroshko OA. 1993. Taxonomic status of the pale martin Riparia diluta (Sharpe and Wyatt, 1893). Russ Ornitol Zh. 2(3):303–323.
- Huang J, Zhou C, Wang LY, Jiang X, Zhang XY, Yue BS, Meng Y. 2019. The complete mitochondrial genome of the Minla cyanouroptera (Passeriformes: Timaliidae). Mitochondrial DNA B Resour. 4(2):3610–3611.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Meklenburtsev RN. 1954. Hirundinidae: 685-750. In: Dementiev GP, Gladkov NA, editors. Birds of the Soviet Union. 6:1–792. Moscow. [In Russian.]
- Shuo L, Chen Y, Liu J-D, Wu Y-H, Xie J-H, Shen Y-W. 2016. Complete mitochondrial genome of Red-rumped Swallow, Cecropis daurica (Passeriformes: Hirundinidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):516–517.
- Saldanha S, Taylor PD, Imlay TL, Leonard ML. 2019. Biological and environmental factors related to communal roosting behavior of breeding Bank Swallow (Riparia riparia). Avian Conserv Ecol. 14(2):21.
- Ye SY, Guo SL, Dou ZL, Wang ZL, Lu JQ. 2016. Nest-site selection of sand martins (Riparia riparia) in the suburbs of Zhengzhou, Henan Province. Acta Ecol Sin. 36(21):7006–7013.
- Zhou YB, Zhang WG, Zhang JX, Zhang J, Hu JC, Yu ZW. 2004. Burrow nest biology of Sand Martin in Nanchong, Sichuan. Chin J Zool. 39(2):66–69.