Abstract
Pareuchiloglanis feae is mainly distributed in Irrawaddy (China and Myanmar), Salween (China and Myanmar), and the Yangtze River drainage areas. In this study, the complete mitochondrial genome of P. feae, which has a circular structure of 16,863 bp, including 13 protein-coding genes, 2 ribosomal RNA (rRNA), 22 transfer RNA (tRNA) genes, and a non-coding control region (D-loop), is being reported for the first time. The A, T, C, and G content were 31.36, 15.69, 28.04, and 24.91%, respectively. Bayesian inference and maximum likelihood method showed that P. feae clustered with P. anteanalis and P. sichuanensis. Our results provide further information and references for clarifying the phylogenetic relationships within Glyptosternoid fishes.
Pareuchiloglanis feae (Vinciguerra, 1890) is a small fish belonging to the order Siluriformes and is distributed in the Irrawaddy (China and Myanmar), Salween (China and Myanmar), and Yangtze River drainage areas (Chu et al. Citation1999; Thomson and Page Citation2006; Fricke et al. Citation2022). This species is usually found in streams around hilly and rocky regions and is commonly known as flat-headed fish. The natural resources of this species have seriously declined in recent years owing to human activities.
In this study, we determined the complete mitochondrial genome sequence of P. feae. Samples were collected from a tributary called Liuchong River (E 105°10′43″, N 26°43′9″), Nayong County, Guizhou Province, that flows into the Wujiang River. A specimen was deposited at the Guizhou Fisheries Research Institute (Sheng Zeng, [email protected]) under the voucher number 20200429001. Total genomic DNA was extracted from the pelvic fin preserved in 95% alcohol using the Qiagen QIAamp tissue kit following the manufacturer’s protocol. The complete mitochondrial genome was sequenced using Illumina NovaSeq. The sequencing results were assembled using SPAdes (Bankevich et al. Citation2012). The structure of the mitochondrial genome was predicted using MITOS (http://mitos.bioinf.uni-leipzig.de) (Bernt et al. Citation2013) and the annotated sequence was archived in GenBank (accession number: MZ901209). The experimental procedures followed in this study complied with the animal welfare and research laws in China and were approved by the Committee of Laboratory Animal Experimentation at the Guizhou Academy of Agriculture Sciences, Guizhou Fisheries Research Institute, China.
The mitochondrial genome of P. feae is a circular molecule of 16,863 bp, which is similar to that of other vertebrates, and includes 13 protein-coding genes, two ribosomal RNA (rRNA), 22 transfer RNA (tRNA) genes, and a non-coding control region (D-loop) (Shao et al. Citation2016; Li et al. Citation2016). The A, T, C, and G content were 31.36, 15.69, 28.04, and 24.91%, respectively. The percentage of A + T was 47.05%, which was lower than that of G + C (52.95%). Except for one protein-coding gene (ND6) and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu and tRNAPro) encoded on the L-strand, all other genes were encoded on the H-strand.
Almost all 13 protein-coding genes started with the typical initiation codon ATG, except for COI and ND3, which started with GTG. Six protein-coding genes terminated with the complete stop codon TAG (ND1, COI, ND5, and ND6) or TAA (ATPase 8 and ND4L), whereas the rest ended with an incomplete stop codon T (ND2, COII, COIII, ND3, ND4, and Cytb) or TA (ATPase6). The D-loop was located between tRNAPho and tRNAPhe (893 bp).
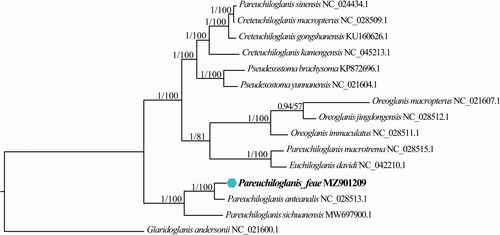
Phylogenetic analyses were performed for mitochondrial genome sequences using the maximum likelihood (ML) method and Bayesian inferences, using MEGA11 (Tamura et al. Citation2021) and MrBayes v3.2.7 (Ronquist et al. Citation2012), respectively. Glaridoglanis andersonii (Siluriformes: Glyptosterninae) was chosen as the outgroup. The topology obtained from Bayesian inferences was similar to that of the ML method. Phylogenetic analysis revealed that P. feae clustered together and formed a sister group to P. anteanalis and P. sichuanensis. However, P. sinensis and P. macrotrem clustered with Creteuchiloglanis macropterus and Euchiloglanis davidi, respectively (). Phylogenetic analyses suggested that the genus Pareuchiloglanis is not monophyletic. Since phylogenetic relationships within Glyptosternoid fishes remain controversial (Peng et al. Citation2004; Yu and He Citation2012; Ma et al. Citation2015), our results may provide more information and references for clarifying the same.
Author contributions
Sheng Zeng was involved in the drafting of the paper, revising it critically for intellectual content. Sheng Zeng and Xue Wang were involved in the conception and design. Wei Liu and Yan Xiang were involved in analysis and interpretation of the data. Lu Zhou was involved in the final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Ethical approval
The experimental procedures followed in this study complied with the animal welfare and research laws in China and were approved by the Committee of Laboratory Animal Experimentation at the Guizhou Academy of Agriculture Sciences, Guizhou Fisheries Research Institute, China.
Supplemental Material
Download MS Word (182 KB)Acknowledgments
Our sincere thanks are given to Wei Wang, Zhuqing Zhang and Wen-Wu Min for helping with fieldwork.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ901209. The associated BioProject, Bio-Sample and SRA numbers are PRJNA757207, SAMN20953560 and SRR15647327 respectively.
Figure 1. Phylogenetic relationships between Pareuchiloglanis feae and 14 other Glyptosternoid fishes based on complete mitochondrial genome sequences. Glaridoglanis andersonii was chosen as the outgroup. Numbers on the nodes are Bayesian posterior probability and bootstrap support for maximum likelihood, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chu X, Cheng B, Dai D. 1999. Faunica Sinica. Class Teleostei, Siluriformes. Beijng: Science Press. [In Chinese, English keys]
- Fricke R, Eschmeyer WN, van der Laan R. 2022. Eschmeyer's catalog of fishes: Genera, species, references. http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp.
- Li B, Tian Z, Qin Y, Hao M, Zhang J. 2016. The complete mitochondrial genome of Pareuchiloglanis gongshanensis (Siluriformes, Sisoridae, Pareuchiloglanis): genome characterization and phylogenetic analysis. Mitochondrial DNA B Resour. 1(1):58–59.
- Ma X, Kang J, Chen W, Zhou C, He S. 2015. Biogeographic history and high-elevation adaptations inferred from the mitochondrial genome of Glyptosternoid fishes (Sisoridae, Siluriformes) from the southeastern Tibetan Plateau. BMC Evol Biol. 15:233.
- Peng Z, He S, Zhang Y. 2004. Phylogenetic relationships of glyptosternoid fishes (Siluriformes: Sisoridae) inferred from mitochondrial cytochrome b gene sequences. Mol Phylogenet Evol. 31(3):979–987.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Shao K, Yan SX, Zhu B, Xu N, Li WT, Xiong MH. 2016. Complete mitochondrial genome of Pareuchiloglanis sinensis (Siluriformes: Sisoridae). Mitochondrial DNA. Part A. Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):713–714.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027.
- Thomson AW, Page L. 2006. Genera of the Asian Catfish families Sisoridae and Erethistidae (Teleostei: Siluriformes). Zootaxa. 1345(1):1–96.
- Yu M, He S. 2012. Phylogenetic relationships and estimation of divergence times among Sisoridae catfishes. Sci China Life Sci. 55(4):312–320.
