Abstract
The complete mitochondrial genome of a Korean endemic species, Iksookimia pacifica (Teleostei: Cypriniformes: Cobitidae) was sequenced using the NGS method. Its total mitogenome was 16,561 bp in length, comprising 13 protein-coding genes (PCGs), 2 ribosomal RNA genes (rRNA), 22 transfer RNA genes (tRNA), and 1 control region (D-loop). The gene order and content were congruent with those of other cobitid species. In the phylogenetic tree using the maximum likelihood method, I. pacifica was clearly distinguished and most closely related to I. koreensis. The mitogenome sequence data of I. pacifica will provide useful information on the phylogenetic relationship among Cobitidae species.
Iksookimia pacifica was previously reported as a synonym for Cobitis tenia granoei (Kim Citation1980) and C. melanoleuca (Nalbant Citation1993), but it was newly classified as C. pacifica based on its body pigmentation and morphological characteristics (Kim et al. Citation1999). However, recently it was reclassified into genus Iksookimia through a taxonomic review of the family Cobitidae by Kim (Citation2009). Iksookimia pacifica is an endemic species and inhabits only the eastern part of the Korean Peninsula, so analyzing their genetic information is very important for conservation biology. Therefore, here we reported the first complete mitochondrial genome of I. pacifica obtained from the fin clip and analyzed its phylogenetic relationship with the other cobitid species.
Specimens of I. pacifica were collected in Goseong, South Korea (38°18′29.35″N, 128°31'40.82″E) in 2021. The specimen was deposited at Environmental Biology Laboratory, Jeonbuk National University in Korea under the voucher number JBNU 39128 (Hyun Tae Kim, [email protected]). For genomic DNA analysis, a piece of pectoral fin was dissected from the specimen and stored in 100% ethyl alcohol. After that, the total DNA was purified with the genomic DNA Prep Kit for blood and tissue (QUIAGEN Co., USA). Sequencing was performed using the Illumina Hi-Seq X-10 platform (San Diego, CA, USA), and the de novo assembler SPAdes 3.13.0 (Bankevich et al. Citation2012) was conducted for mitogenome construction. The mitogenome sequence was annotated using the MITOS web server (Bernt et al. Citation2013).
The complete mitogenome of I. pacifica (OM312055) is composed 16,561 bp of nucleotide. The genome consists of 13 PCGs, 22 tRNA genes, 2 rRNA genes, and 1 control region (D-loop). The gene order and arrangement were completely identical with the other cobitid species. The most common start codon in all PCGs is 'ATG', with the exception of COX1 that start with 'GTG'. For termination codon, seven PCGs (ND1, COX1, ATP8, ATP6, ND4L, ND5, ND6) have a complete ‘TAA’. But five PCGs (ND2, COX2, COX3, ND3, CYTB) have ‘T—’, an incomplete terminal codon, and only ND4 had ‘TA–’. The overall nucleotide compositions were 29.3% A, 27.5% T, 26.1% C, and 17.1% G, respectively.
The dataset for molecular phylogenetic analysis to understand the taxonomic relationship between I. pacifica (OM312055) and other sister species included 13 PCGs and 2 rRNA sequences of 16 other cobitid species, two families belonging to order Cypriniformes, and one species of Siluridae was added as an outgroup (). For maximum likelihood analysis, the dataset was analyzed using MEGA version X (Kumar et al. Citation2018) with GTR + G + I model (Nei and Kumar Citation2000). The bootstrap resampling was accomplished with 1,000 iterations.
Figure 1. Phylogenetic tree of maximum likelihood (ML) method based on the nucleotide sequences of 13 PCGs and 2 rRNAs of 17 Cobitid species, including I. pacifica (OM312055), two families belonging to order Cypriniformes, and one Siluriformes species. Bootstrap support values based on 1,000 replicates are displayed on each node as >70.
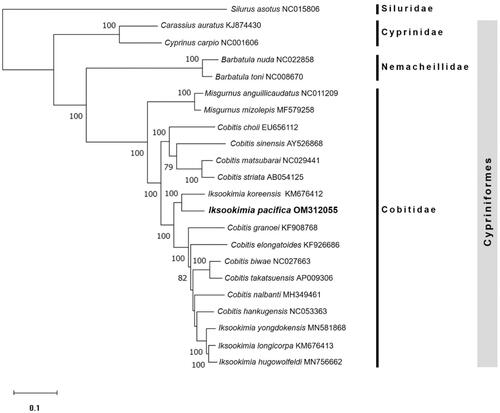
In the phylogenetic analysis, I. pacifica was clearly distinguished from other species and was most closely related to I. koreensis (). Our result suggests that I. pacifica and I. koreensis seem to have differentiated from the same recent common ancestor, and it was supported by the research that their habitats were interconnected before the geographical uplift event that occurred on the Korean Peninsula during the Miocene (Kwan et al. Citation2018). And it was also confirmed that Cobitidae species form a distinct monophyletic clade within the order Cypriniformes. On the other hand, I. pacifica and I. koreensis did not form a group with other Iksookimia species, consistent with other studies (Park et al. Citation2018; Kim et al. Citation2020). Therefore, a taxonomic review of these discrepancies is necessary, and our results are expected to provide useful information for reconstructing the phylogenetic relationship between Cobitidae species in future studies.
Author contributions
SWY and HTK conceived and designed the experiments. SWY and HTK collected and identified the samples. SWY and HTK performed the experiments, analyzed the data, and wrote the manuscript. SWY and HTK prepared the figures. All authors reviewed the manuscript.
Ethical approval
All of the fish in this study were treated based on the ‘Ethical review process by Jeonbuk National University Institutional Animal Care and Use Committee (JBNUIACUC).’
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession number OM312055. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA798219, SRR 17652036, and SAMN 25050457, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Kim IS, Park JY, Nalbant TT. 1999. The far-east species of the genus Cobitis with description of three new taxa (Pisces; Ostairophysi; Cobitidae). Trav Mus Nat Hist Nat ‘Griore Antipa. 44:373–391.
- Kim IS. 1980. Systematic studies of on the fishes of the family Cobitidae (Order Cypriniformes) in Korea. 1. Three unrecoreded species and subspecies of the genus Cobitis from Korea. Korean J Zool. 23:239–250.
- Kim IS. 2009. A review of the spined loaches, family Cobitidae (Cypriniformes) in Korea. Korean J Ichthyol. 21:7–28.
- Kim PJ, Han JH, An SL. 2020. Complete mitochondrial genome of Korean endemic species, Iksookimia yongdokensis (Actinopterygii, Cypriniformes, Cobitidae). Mitochondr DNA B Resour. 5(1):633–634.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Kwan YS, Kim D, Ko M, Lee W, Won Y. 2018. Multi-locus phylogenetic analyses support the monophyly and the Miocene diversification of Iksookimia (Teleostei: Cypriniformes: Cobitidae). Syst Biodiver. 16(1):81–88.
- Nalbant TT. 1993. Some problems in the systematics of the genus Cobitis and its relative (Pisces: Ostariophysi, Cobitidae). Rev Roum Biol. 38:101–110.
- Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York.
- Park HK, Kim KS, Kim KY, Bang IC. 2018. The full-length mitochondrial genome of Cobitis nalbanti (Teleostei: Cypriniformes: Cobitidae). Mitochondr DNA B Resour. 3(2):870–871.
