Abstract
In this study, the complete mitochondrial genome (mitogenome) of Sinularia humilis van Ofwegen, 2008 was determined using next-generation sequencing (NGS) method. The mitogenome of S. humilis is 18,743 bp in length, containing 14 protein-coding genes (PCGs), two ribosomal RNAs (rRNA) and one tRNA (tRNA-Met), which has same gene order with other species of Sinularia. ATG was determined as start codon in all 14 PCGs. Eight TAG, five TAA, and one incomplete codons (T-) were found as stop codon. Phylogenetic analysis of the small number of available mitogenomes showed that S. humilis is closely related to Sinularia ceramensis and Sinularia peculiaris.
The genus Sinularia with approximately 170–190 biological species is a large group in Octocorallia, commonly distributed in tropical zones of shallow water to deep reefs (van Ofwegen Citation2000; Fabricius and Alderslade Citation2001; McFadden et al. Citation2009). van Ofwegen has described species of Sinularia humilis from Palau (Micronesia) in 2008. Further studies have documented distribution of this species in South China Sea (Benayahu et al. Citation2012, Citation2018).
So far, only six mitogenomes of genus Sinularia have been sequenced (Asem et al. Citation2019; Chen et al. Citation2019; Shen et al. Citation2021); their mitogenome lengths ranged from 18,730 to 18,742 bp. In the present study, complete mitochondrial genome of S. humilis (GenBank no. OK641586) was sequenced and annotated to study the mitogenomic characteristics and its phylogenetic relationships within Sinularia.
With regard to regulations of Department of Science and Technology of Hainan province government, a permission was obtained to collect a sample from China South Sea following R&D program under ZDKJ2019011-03-02 reference number. A specimen of S. humilis was collected from the South China Sea (Small Island, Sanya, China; 18°15′12″N-109°30′13″E) and stored in Hainan Tropical Ocean University Museum of Zoology (specimen voucher: HTOU-SiHu001; Chaojie Yang; [email protected]). The specimen was identified based on morphology of sclerites (Benayahu, personal communication). Total genomic DNA was extracted with Genomic DNA Isolation Kit no. B518221 (Sangon Biotech Co., Shanghai, China) (Asem et al. Citation2021).
The quality of extracted DNA was checked on a 1.5% agarose gel and then quantified using a Micro-volume Spectrophotometer (MaestroGen Inc., Hsinchu City, Taiwan). An amount of 600 ng of total DNA was used to construct the genomic library with paired-end (2 × 150 bp) followed by next-generation sequencing (NGS) (10 Gb) approach. The sequencing was performed on the Illumina HiSeq X-ten sequencing platform (Novogene Co., Tianjin, China). Total sequencing included effective rate, 0.03% of error rate and 90.31% of Q30. Quality of reads were controlled by FastQC. De novo assemblies were performed with Geneious 9.1 utilizing the mitogenome of Sinularia ceramensis (GenBank no. NC_044122) as reference map with default parameter settings (Kearse et al. Citation2012). The position of tRNA-Met gene was determined by ARWEN software (http://130.235.46.10/ARWEN/). PCGs and rRNAs were annotated by multiple sequence alignments to the reference mitogenome using BioEdit program (Hall Citation1999). In addition, all PCGs were translated into amino acids by the ExPASy online program (https://web.expasy.org/translate/) and sequences were examined to ensure that each could encode a functional protein.
The length of mitogenome of S. humilis was 18,743 bp including 14 protein-coding genes (PCGs: 14,804 bp), two ribosomal RNAs (rRNAs: 3260 bp), and one transfer RNA (tRNA-Met: 17 bp). The overall nucleotide composition of the heavy strand was as follows: 30.35% A, 16.52% C, 19.29% G, and 33.83% T, with a total A + T content of 64.18%. Four PCGs including COX3, ATP6, ATP8, and COX2 and tRNA-Met were located on the light strand. All PCGs encoded with common ATG start codon. Stop codons included eight TAG (ND1, CYTB, ND6, ND3, ND2, ND5, COXIII, and COXII), five TAA (ND4L, MutS, ND4, ATP6, and ATP8), and a non-complete codon T (COXI). The 12S rRNA and 16S rRNA were encoded on the heavy strand from 1589 to 2634 (1046 bp) and 9159 to 11,369 (2211 bp), respectively. Same as other Sinularia mitogenome, 16S rRNA showed a rather higher A + T content (58.07% vs. 56.69%) in S. humilis. The longest overlap and gap were observed between ND2/ND5 with 13 bp and COX2/COX1 with 112 bp, respectively.
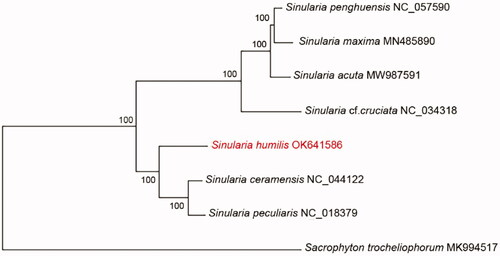
The phylogenetic relationship of S. humilis among other species of Sinularia was inferred with a concatenated dataset including the 14 PCGs and two rRNAs using maximum-likelihood (ML). Sarcophyton trocheliophorum (MK994517) was used as an outgroup (Shen et al. Citation2019). The ML phylogenetic analysis was performed using MEGA X. The best-fitting nucleotide substitution model was calculated based on the results of the MrModelTest 2.2 (Alfaro et al. Citation2003) and HKY + G was chosen as the best-fit model with 1000 bootstrap replicates (). Regarding available mitogenomes, results revealed that Sinularia was divided in two clades and that S. humilis was closely related to Sinularia ceramensis and Sinularia peculiaris. Further studies with large number of Sinularia mitogenomes need to represent clear status of species of genus Sinularia.
Author contributions
Ch. Y. designed the research. Material preparation, data collection and analysis, and interpretation of the data were performed by Ch. Y. Molecular analysis and the final draft of the manuscript was revised by Zh. W. and Y. H. All authors commented on previous versions of the manuscript. All authors have read and agreed to the published version of the manuscript.
Acknowledgements
We thank Dr. Chengyue Liu and Dr. Zhi Chen for their help to collect samples and revise the manuscript, respectively. The authors thank Prof. Yehuda Benayahu for his contribution to sample identification.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number OK641586. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA780662, SAMN23175337, and SRR16960927, respectively.
Additional information
Funding
References
- Alfaro ME, Stefan Z, François L. 2003. Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol Biol Evol. 20(2):255–266.
- Asem A, Eimanifar A, Li W, Shen C-Y, Shikhsarmast FM, Dan Y-T, Lu H, Zhou Y, Chen Y, Wang P-Z, et al. 2021. Reanalysis and revision of the complete mitochondrial genome of Artemia urmiana Günther, 1899 (Crustacea: Anostraca). Diversity. 13(1):14.
- Asem A, Lu H, Wang P-Z, Li W. 2019. The complete mitochondrial genome of Sinularia ceramensis Verseveldt, 1977 (Octocorallia: Alcyonacea) using next-generation sequencing. Mitochondrial DNA B Resour. 4(1):815–816.
- Benayahu Y, Ofwegen L, Dai CF, Jeng M-S, Soong K, Shlagman A, Hsieh HJ, McFadden CS. 2012. Diversity, distribution, and molecular systematics of Octocorals (Coelenterata: Anthozoa) of the Penghu Archipelago, Taiwan. Zool Stud. 51(8):1529–1548.
- Benayahu Y, van Ofwegen LP, Dai C-F, Jeng M-S, Soong K, Shlagman A, Du SW, Hong P, Imam NH, Chung A, et al. 2018. The Octocorals of Dongsha Atoll (South China Sea): an iterative approach to species identification using classical taxonomy and molecular barcodes. Zool Stud. 57:e50.
- Chen Y, Dan Y-T, Lu H, Wang P-Z, Asem A, Li W. 2019. The complete mitochondrial genome of Sinularia maxima Verseveldt, 1971 (Octocorallia: Alcyonacea) using next-generation sequencing. Mitochondrial DNA B Resour. 4(2):3425–3426.
- Fabricius KE, Alderslade P. 2001. Soft corals and sea fans: a comprehensive guide to the tropical shallow water genera of the central-west Pacific, the Indian Ocean and the Red Sea. Townsville: Australian Institute of Marine Science.
- Hall TA. 1999. Bio Edit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. 41(41):95–98.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- McFadden CS, van Ofwegen LP, Beckman EJ, Benayahu Y, Alderslade P. 2009. Molecular systematics of the speciose Indo-Pacific soft coral genus, Sinularia (Anthozoa: Octocorallia). Invert Biol. 128(4):303–323.
- Shen C-Y, Dan Y-T, Asem A, Wang P-Z, Xue W, Tong X-B, Li W. 2019. The complete mitochondrial genome of soft coral Sarcophyton trocheliophorum (Cnidaria: Anthozoa) using next-generation sequencing. Mitochondrial DNA B Resour. 4(2):3734–3735.
- Shen C-Y, Wang P, Zh Xue W, Liu Zh H, Zhao J-Y, Tong X-B, Liu Ch W, Wu X-F, Mao X-N, Tian S-H, et al. 2021. The complete mitochondrial genome of soft coral Sinularia penghuensis Ofwegen and Benayahu, 2012 (Octocorallia: Alcyonacea): the analysis of mitogenome organization and phylogeny. Mitochondrial DNA B Resour. 6(4):1348–1350.
- van Ofwegen LP. 2000. Status of knowledge of the Indo-Pacific soft coral genus Sinularia May, 1898 (Anthozoa: Octocorallia). Proceedings 9th International Coral Reef Symposium. p. 167–171.