Abstract
Morphostenophanes yunnanus (Zhou, Citation2020) is widely distributed from central to eastern Yunnan with distinct geographical variations in morphology. Beetles were collected in Manwan Town, and a mitochondrial genome sequence (GenBank accession number MZ298928) of this species was sequenced using the MGI-SEQ 2000 platform, assembled using NOVOPlasty v4.3.1, and characterized. The mitogenome was a circular DNA molecule of 15,690 bp with 64.710% AT content, which comprised 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and one control region. The protein-coding genes showed the typical ATN (Met) and TTG (Met) start codons, except nad1 and cox1 (TTG as start codon), and were terminated by typical TAN stop codons. The maximum-likelihood polygenetic tree was generated using protein sequences of thirteen protein-coding regions of seventeen mitogenomes with mtREV + G + F + I with 1000 replicates under the Bayesian information criterion using MEGA 11, which showed that M. yunnanus was the most closely related to M. sinicus. This study provides essential genetic and molecular data for phylogenetic analyses of the genus Morphostenophanes.
Morphostenophanes yunnanus (Zhou, Citation2020) is widely distributed from central to eastern Yunnan with distinct geographical variations in morphology (Zhou Citation2020). It has gray-black shagreened mouth parts and dark-brown tarsi (Zhou Citation2020). M. yunnanus can be divided into seven geographical populations (Zhou Citation2020). The complete mitochondrial genome of M. yunnanus of the Manwan Town population was sequenced and characterized.
Adult M. yunnanus specimens were collected from Shuibatou Village (100.32963° N, 24.68407° E), Manwan Town, Yun County, Lincang City, Yunnan Province, China, on 24 September 2020, and deposited in the Insect Collection of Institute of Plant Protection, Guangxi Academy of Agricultural Sciences (http://www.gxaas.net/s.php/zwbhyjs/, Xuyuan Gao, [email protected]) under the voucher number GIPP-20200924-001. Genomic DNA was isolated and subjected to paired-end sequencing (2 × 150 bp) of 400 bp inserts using the MGI-SEQ 2000 platform. We produced ∼11.537 Gb of raw data, of which 11.301 Gb (97.76%) were clean high-quality data using SOAPnuke version 2.1.0 (Chen et al. Citation2018) with default parameters. The genome was assembled de novo using NOVOPlasty v4.3.1 (Dierckxsens et al. Citation2017) with default parameters and the mitogenome of Morphostenophanes sinicus (MW853764.1) (Bai, Gao, et al. Citation2021) as a seed sequence (no extending the seed directly). Three pairs of primers were designed corresponding to the cox1, nad5, and control regions (CR, also an AT-rich region) to verify the accuracy of the genome assembly. PCR products were sequenced from other M. yunnanus specimens using Sanger sequencing using an ABI 3730 automatic sequencer and assembled manually (Supplementary 1, 2), and only a few bases were found to be different (Supplementary 1).
The circular mitogenome (nucleotide composition: 37.935% A, 26.775% T, 23.391% C, and 11.899% G; and 64.710% AT content) of M. yunnanus (MZ298928.1) was 15,690 bp in length. Using Perna and Kocher’s formula (Perna and Kocher Citation1995), The AT and GC skews of the major strand of the mitogenome were estimated to be 0.172 and −0.326, respectively. MITOS (http://mitos.bioinf.uni-leipzig.de/) (Bernt et al. Citation2013) was used for the sequence annotation, revealing 13 protein-coding genes (PCGs), 1 CR, 22 tRNA genes, and two rRNA genes. Using the genomes of Promethis valgipes valgipes (MW201671.1) (Bai, Chen, et al. Citation2021), Tenebrio obscurus (MG739327.1) (Bai et al. Citation2018), M. sinicus (MW853764.1) (Bai, Gao, et al. Citation2021), Blaps rhynchoptera (MN267802.1) (Yang et al. Citation2019), and Zophobas atratus (MK140669.1) (Bai et al. Citation2019) as a reference, the start and stop codons of PCGs were corrected manually. All 13 PCGs had traditional ATN (Met) start codons, except for nad1 and cox1 (TTG): atp8 starts with an ATC start codon; two PCGs (nad3 and nad6) start with an ATA start codon; five PCGs (atp6, cox3, nad4, nad4l, and cob) start with an ATG start codon; three PCGs (nad2, cox2, and nad5) start with an ATT start codon. All 13 PCGs using traditional TAN as stop codons: six PCGs (nad2, cox1, nad4, atp6, nad6, and cob) ended with a TAA stop codon; five PCGs (cox2, atp8, nad3, nad4l, and nad1) ended with a TAG stop codon; two PCGs (cox3 and nad5) had an incomplete stop codon (T), consisting of a codon that was completed by the addition of A nucleotides at the 3′ end of the encoded mRNA. The 22 tRNA ranged from 60 (tRNA-Cys and tRNA-Gly) to 72 bp (tRNA-His). The lrRNA and srRNA were 1195 and 756 bp in length, respectively. The CR of the mitogenome was 1121 bp with a 79.572% AT content, which was located between the srRNA and tRNA-Ile genes.
For phylogenetic analyses, M. yunnanus mitochondrial PCGs and 16 other insect species were used to construct a maximum-likelihood (ML) phylogenetic tree. Protein sequences on thirteen protein-coding regions of their mitogenomes were aligned using MEGA 11 software (Tamura et al. Citation2021) with MUSCLE program (Edgar Citation2004) with default specifies. The ML Model with the lowest Bayesian Information Criterion (BIC) scores is considered to be the best. According to BIC (=93859.54), General Reversible Mitochondrial (mtREV24) + a discrete Gamma distribution (G) + amino acid frequencies (F) + evolutionarily invariable (I) with 1000 replicates was chosen to construct a phylogenetic tree using MEGA 11 (Tamura et al. Citation2021) (). Sixteen mitogenomes of species belonging to the family Tenebrionidae and the mitogenome of Lepisma saccharina (Bai et al. Citation2020) were selected as the outgroup. The resulting polygenetic tree revealed that M. yunnanus was the most closely related to M. sinicus. Overall, our study provides insights into the mitogenome of M. yunnanus and essential genetic and molecular data for phylogenetic analyses of the species belonging to the genus Morphostenophanes.
Figure 1. Maximum-likelihood phylogenetic tree of Morphostenophanes yunnanus and those of 16 other insect species based on protein sequences of thirteen protein-coding regions of their mitogenomes.
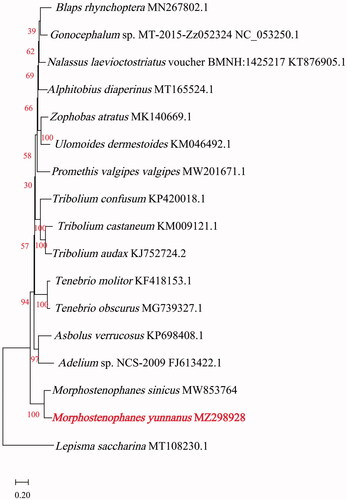
Phylogenetic relationships were inferred using maximum likelihood and mtREV + G (parameter = 0.4735) + F + I (17.50% sites). The highest log-likelihood of the tree is −48283.84. The percentage of trees is shown below the branches in red. The complete mitogenome of M. yunnanus (GenBank accession number MZ298928) determined in this study is indicated in red.
Author contributions
Yu Bai analyzed the data, uploaded the analysis data, involved in certain tools for analysis, drafted the paper, and approved the final draft. Yang Kang performed the experiments. Lin Ye collected and analyzed data. Xuyuan Gao identified insects, contributed reagents/materials, involved in the conception and design of the work, performed the experiments, prepared figure, and approved and published the final draft. All authors agree to be accountable for all aspects of the work.
Ethical approval
This research does not involve ethical research. Insects are invertebrates, and there are no ethics involved in using them in experiments.
Supplemental Material
Download MS Word (20.9 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MZ298928. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA729642, SRR14523735, and SAMN19159615, respectively.
Additional information
Funding
References
- Bai Y, Chen J, Li G, Luo J, Wang H, Yang Y, Liang S, Ouyang B. 2021. Complete mitochondrial genome of Promethis valgipes valgipes (Marseul) (Insecta: Coleoptera: Tenebrionidae). Mitochondrial DNA B Resour. 6(2):538–539.
- Bai Y, Chen J, Li G, Wang H, Luo J, Li C. 2020. Complete mitochondrial genome of the common silverfish Lepisma saccharina (Insecta: Zygentoma: Lepismatidae). Mitochondrial DNA B. 5(2):1552–1553.
- Bai Y, Gao X, Yu Y, Long X, Zeng X, Wei D, Ye L. 2021. Complete mitochondrial genome of Morphostenophanes sinicus (Zhou, 2020) (Insecta: Coleoptera: Tenebrionidae). Mitochondrial DNA B Resour. 6(10):2946–2948.
- Bai Y, Li C, Yang M, Liang S. 2018. Complete mitochondrial genome of the dark mealworm Tenebrio obscurus Fabricius (Insecta: Coleoptera: Tenebrionidae). Mitochondrial DNA B Resour. 3(1):171–172.
- Bai Y, Wang H, Li G, Luo J, Liang S, Li C. 2019. Complete mitochondrial genome of the super mealworm Zophobas atratus (Fab.) (Insecta: Coleoptera: Tenebrionidae). Mitochondrial DNA B. 4(1):1300–1301.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, Li S, Li Y, Ye J, Yu C, Li Z, et al. 2018. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 7(1):1–6.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- Perna NT, Kocher TD. 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 41(3):353–358.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027.
- Yang Y, Bai Y, Zheng J, Chen J, Ouyang B, Liang S. 2019. Characterization of the complete mitochondrial genome of Blaps rynchopetera Fairmaire (Insecta: Coleoptera: Tenebrionidae) from Dali. Mitochondrial DNA B Resour. 4(2):3167–3168.
- Zhou D. 2020. A revision of the genus Morphostenophanes Pic, 1925 (Coleoptera, Tenebrionidae, Stenochiinae, Cnodalonini). Zootaxa. 4769(1):zootaxa.4769.1.1.
