Abstract
The complete mitogenome sequence of Coilia brachygnathus (Kreyenberg & Pappenheim, 1908) from Wabu Lake in Huai River Basin was annotated and characterized in this study. This mitochondrial genome is a circular DNA molecule of 16.896 bp in size with 57.52% AT content, including 13 protein-coding genes (PCGs), two ribosomal RNA genes (rRNAs), 22 transfer RNA genes (tRNAs), and an AT-rich region (control region) as other bony fishes. There are a total of 10 overlap locations and 15 intergenic spacer regions throughout the mitogenome of C. brachygnathus. All PCGs employed a standard ATG as a start codon, except cytochrome c oxidase 1 (cox1) with GTG. In addition, TAA or TAG was identified as the typical stop codon. A phylogenetic tree reconstructed with the maximum likelihood method depicted a clone relationship with eight species of genus Coilia and our previous study based on the amino acid sequences of 13 mitochondrial PCGs. The complete mitochondrial genome is a valuable resource in classifying and conserving C. brachygnathus.
Coilia brachygnathus (Clupeiformes: Engraulidae: Coilinae), known as the only freshwater fish in the genus Coilia, is a kind of small fish growing, developing, and reproducing in Yangtze River and its affiliated water bodies (Whitehead Citation1985). The Huai River which flows into the Yangtze River at the lower reaches of Hongze Lake is known as one of the affiliated water bodies. It was shown by previous investigations that these species were abundant in the Huai River. This species is an important food source for local populations, as well as a bait for large carnivorous fish, C. brachygnathus plays an important role in the food chain in the Huai River (Wang et al. Citation2020). It is acknowledged that under the influence of various factors, such as overfishing, environmental pollution, and habitat destruction, the wild resources of C. brachygnathus were under great threat. Till now, although the multiple genomes of C. brachygnathus from the Huai River have been published, the populations they sampled belonged to the migratory ones in the mainstream of the Huai River rather than the land-locked population, while the populations living in the Wabu Lake were typical land-locked (Hu et al. Citation2019). There is no denying the fact that our achievement will facilitate further studies on Coilia fishes, including the phylogenetic relationship between the species and population structure of C. brachygnathus from the Huai River.
The complete mitochondrial genome of C. brachygnathus was sequenced and characterized in this study. The samples of C. brachygnathus were collected from the Wabu Lake (E116.9076 and N32.3861) in Huainan City, Anhui Province, China, and stored in the Aquatic Museum of Xinyang Agriculture and Forestry University (https://www.xyafu.edu.cn/, Liangjie Zhao and [email protected]) under the voucher number XYAFU-Mu-1803226. Before further processing, the samples were placed in 100% ethanol in the process of collection and were stored at ambient temperature (−80 °C) until the extraction of DNA. The extraction of total DNA from the partial body tissue was carried out employing the TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, China). With the quality of the separated DNA detected by 1.5% electrophoresis, the DNA was stored at ambient temperature (−20 °C) until the complete mitogenomes were amplified by PCR. Long and accurate PCR (LA PCR) was performed to amplify the complete mitochondrial genome sequence using a set of specific primers. Sequencing was conducted using the primer walking method on an ABI 3730XL DNA Analyzer (Applied Biosystems Inc., Foster City, CA, USA).
With a length of 16.896 bp, the complete mitochondrial genome of C. brachygnathus has been deposited in GenBank with accession No. ON209111. It is composed of 13 protein-coding genes, 22 tRNA genes, and two rRNA genes. The encoding genes of mitogenome were located on H-strand except for nad6 and eight tRNA genes, which were transcribed from L-strand. With only cox1 starting with GTG, the ATG and GTG are used as the start codon. Except for cox2, nad4, and Cytb with an incomplete stop codon ‘T–,’ the remaining protein-coding genes stop with TAG or TAA. The tRNA genes were interspersed among the mitochondrial genome, with the sizes ranging from 66 to 75 bp, and it was found that twenty-two tRNA genes were folded into a typical cloverleaf secondary structure (Donath et al. Citation2019), which was similar to other Coilia mitochondrial genomes (Hu et al. Citation2019; Yang et al. Citation2019). It was estimated that the overall basic composition of the heavy strand in C. brachygnathus was 31.21% for A, 26.30% for T, 15.47% for G, and 27.01% for C, with an AT content of 57.52%, respectively. As a result, it indicated that there was an obvious antiguanine bias. The 13 PCGs have a size of 11.424 bp in total, accounting for 67.61% of the whole mitogenome.
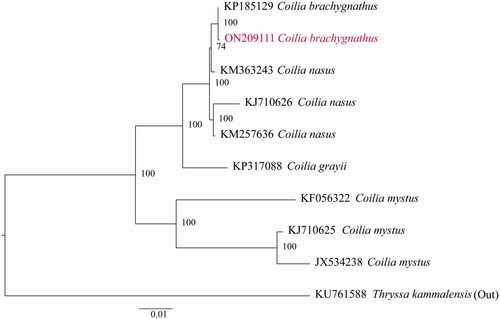
We selected eight complete mitogenome data of Coilia obtained from the GenBank database. In addition, Thryssa kammalensis (Bleeker, 1849) (KU761588) was deemed as an outgroup. Apart from that, PartitionFinder 2.1.1 was employed to select the optimal evolutionary models for phylogenetic analysis (Lanfear et al. Citation2017). The preparation for the amino acid sequence of the 13 genes for all the selected species was conducted in the following specific order: nad1, nad2, cox1, cox2, atp8, atp6, cox3, nad3, nad4L, nad4, nad5, nad6, and cob. Phylogenetic analysis was carried out using the IQ-TREE v1.6.10 (Minh et al. Citation2020). It was shown by the results () that the sample from Wabu Lake should be classified as C. brachygnathus, and it is a member of the genus Coilia.
Author contributions
Liangjie Zhao conceived and designed the experiments. Tiezhu Yang, Yuan Tian, Shijie Yang, and Leping Wang performed the experiments. Tiezhu Yang and Shijie Yang analyzed the data. Yuan Tian and Tiezhu Yang wrote the paper. Liangjie Zhao and Leping Wang helped to proofread the paper. All authors have read and agreed to the published version of the manuscript.
Ethical approval
Experiments were performed in accordance with the recommendations of the Ethics Committee of Xinyang Agriculture and Forestry University. These policies were enacted according to the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. ON209111.
Additional information
Funding
References
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552.
- Hu Y, Duan G, Ling J, Jiang H. 2019. The complete mitogenome and phylogenetic analysis of Coilia brachygnathus (Clupeiformes: Engraulidae) from Chaohu Lake. Mitochondrial DNA B. 4(1):1317–1318.
- Hu Y, Duan G, Zhou H, Pan T, Ling J, Jiang H. 2019. The complete mitochondrial genome of Coilia brachygnathus (Clupeiformes: Engraulidae: Coilinae) from Huaihe river. Mitochondrial DNA B. 4(1):618–619.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Wang H, Hu Y, Jiang H, Duan G, Ling J, Pan T, Zhou H, Chen X, Hou G. 2020. The genetic diversity of Coilia brachygnathus in the Huaihe River basin of Anhui province. Chinese J Ecol. 55:8.
- Whitehead PJ. 1985. FAO species catalogue. Clupeoid fishes of the world (suborder Clupeoidei). Part 1-Chirocentridae, Clupeidae and Pristigasteridae. FAO Fish Synop. 125(7):1–303.
- Yang F, Tao Y, Duan B, Zhang Q, Zhai X, Li Y. 2019. Complete mitochondrial genome of the Yangtze grenadier anchovy, Coilia brachygnathus (Clupeiformes: Engraulidae) from the upper Yangtze River. Mitochondrial DNA B. 4(1):1140–1141.