Abstract
Species belonging to Ulva (Ulvophyceae, Chlorophyta) are one of the major members of invasive seaweeds. Ulva californica Wille 1899 was originally believed to be native to the Pacific coast of North America, while in recent years it has been reported as exotic species, or new record, in Europe, the Mediterranean, Asia, and Oceania. However, the paths of global dispersal of U. californica are unclear. In addition, the species boundary between U. californica and a related species is somewhat disputed. Here, we reported that the complete chloroplast genome of U. californica is 92,126 bp in size, harboring 96 genes (GenBank accession no. MZ561475). The overall base composition was A (37.9%), T (37.4%), C (12.3%), and G (12.4%), similar to those from other Ulva species. The phylogenomic analysis showed that although U. californica was genetically closer to Ulva aragoënsis (Bliding) Maggs 2018 in [Krupnik N et al., 2018], they were clearly distinguishable, supporting the recent opinion that they should be separated into different species. The chloroplast genome data of U. californica would provide plenty resources for phylogeography analysis and monitor on bioinvasion.
Due to global warming and a variety of anthropogenic activities such as mariculture and discharge of ballast water, macroalgae can spread or be introduced into new habitats, and species in the genus Ulva (Ulvophyceae, Chlorophyta) are one of the major members (Verlaque and Breton Citation2019; Xie et al. Citation2020; Liu et al. Citation2022). Ulva californica Wille 1899 was initially described with the type location at La Jolla, California (Collins et al. Citation1899), and two other taxa, i.e. U. angusta Setchell & N.L. Gardner 1920 and U. scagelii Chihara 1969, were later placed into the synonymy with it (Tanner Citation1986). U. californica was originally believed to be native to the Pacific coast of North America (Scagel et al. Citation1989; Wolf et al. Citation2012), but some speculate that it may be more widespread (Loughnane et al. Citation2008). Nevertheless, in recent years U. californica has been reported as exotic species, or new record, in Europe (Hayden and Waaland Citation2004; Loughnane et al. Citation2008), the Mediterranean (Wolf et al. Citation2012; Sfriso et al. Citation2020), Asia (Kawai et al. Citation2007), and Oceania (Heesch et al. Citation2009; Kirkendale et al. Citation2013), even rapid local spreads after introduction have been observed in Germany (Steinhagen et al. Citation2019), and China (Wei et al. Citation2022). However, the paths of global dispersal of U. californica are unclear. In addition, the species boundary between U. californica and a related species is somewhat disputed. According to results of molecular identification and hybridization examination, U. mediterranea Alongi, Cormaci & G.Furnari 2014, which was later revised to U. aragoënsis (Bliding) Maggs 2018 in [Krupnik N et al., 2018] (Krupnik et al. Citation2018), was distinguished from U. californica and U. flexuosa Wulfen 1803 (Hiraoka et al. Citation2017), while in a later study these species were still combined into one complex (Steinhagen et al. Citation2019). Organelle genome data from U. californica can help clarify this controversy, and provide sufficient molecular markers to reveal geographic origins and dispersal routes.
Here, we sequenced the chloroplast genome of U. californica sample U484-3, which was collected from Putian, Fujian Province, China in 2021 (25°12′17″N, 119°33′51″E), and cultured in laboratory with Von Stosch’s Enriched (VSE) medium at 16 °C under a light intensity of 80–100 μmol·m−2·s−1. This kind of plant study did not need specific permissions from the ethics committee of Institute of Oceanology, Chinese Academy of Sciences (IOCAS), and the field collection was carried out following the National standards of the People's Republic of China (Citation2007). A specimen was deposited in Marine Biological Museum of Chinese Academy of Sciences (MBMCAS) at IOCAS (http://www.qdio.cas.cn, Yongqiang Wang, [email protected]) under the voucher number MBM287040.
The algal tissue was sent to BENAGEN Co. Ltd. (Wuhan, China) for high-throughput sequencing. Total genomic DNA was extracted using a Plant Genomic DNA Extraction Kit (Tiangen Biotech Co., Ltd., Beijing, China). The library of genomic DNA was sequenced using the Illumina and Nanopore platform. The read length for Illumina was 150 bp. The total amount and base of reads were 30,479,638 and 4.6 Gbp for Illumina, and 1,149,830 and 5.4 Gbp for Nanopore platform, respectively. A short sequence assembly software Flye v.2.8.3 was used to assemble clean data (Kolmogorov et al. Citation2020) and the obtained complete chloroplast genome sequence was annotated with PGA (Qu et al. Citation2019).
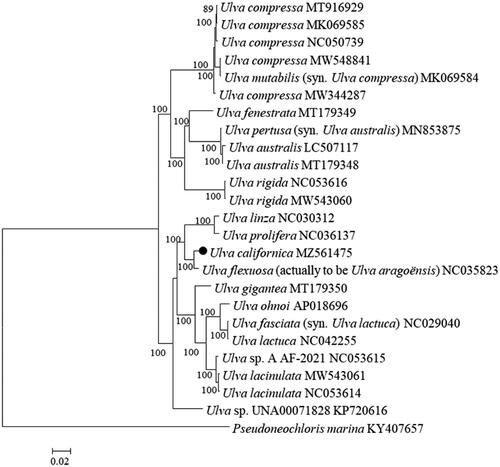
The complete chloroplast genome of U. californica was 92,126 bp in size (GenBank accession no. MZ561475). The overall base composition was A (37.9%), T (37.4%), C (12.3%), G (12.4%), and the percentage of AT (75.3%) is much higher than CG (24.7%), which were similar to those from other Ulva species. This genome encodes 96 genes, including 68 protein-coding genes, 26 transfer RNAs genes, and two ribosomal RNAs genes. Using MEGA 6.0 with a GTR + G+I model (Tamura et al. Citation2013), a maximum-likelihood (ML) phylogenetic tree was constructed with 24 complete chloroplast genomes of Ulva and one chloroplast genome of Pseudoneochloris marina as the outgroup. It was shown that, although U. californica was genetically closer to Ulva aragoënsis (Cai et al. Citation2017), they were clearly distinguishable (), supporting the recent opinion that they should be separated into different species (Hiraoka et al. Citation2017). The data of U. californica chloroplast genomes can be used as resources for phylogeography analysis and monitor on bioinvasion, even risk of green tides dominated by this species (Bae Citation2010).
Figure 1. Phylogenetic tree based on maximum-likelihood (ML) analysis with 24 Ulva chloroplast genomes and one chloroplast genome from Pseudoneochloris marina as the outgroup. Numbers above each node indicate the bootstrap support value. The black dot represents the sequence from the sample used in this study.
Author contributions
Conception and design, P. Jiang; sampling, X. Lin and X. Wei; analysis and interpretation of the data, X. Lin and W. Liu; drafting of the paper, X. Lin and W. Liu; revising it critically for intellectual content, P. Jiang; final approval of the version to be published, P. Jiang. All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MZ561475. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA788066, SRR17206483, and SAMN23896768, respectively.
Additional information
Funding
References
- Bae EH. 2010. Ulotrichales, Ulvales. In: Bae EH, Kim HS, Kwon CJ, Hwang IK, Kim GH, Klochkova TA, editors. Algal flora of Korea. Incheon, Korea: National Institute of Biological Resources; p. 7–52.
- Cai CE, Wang LK, Zhou LJ, He PM, Jiao BH. 2017. Complete chloroplast genome of green tide algae Ulva flexuosa (Ulvophyceae, Chlorophyta) with comparative analysis. PLOS One. 12(9):e0184196.
- Collins FS, Holden I, Setchell WA. 1899. Phycotheca Boreali-Americana. A collection of dried specimens of the algae of North America. Malden (MA).
- Hayden HS, Waaland JR. 2004. A molecular systematic study of Ulva (Ulvaceae, Ulvales) from the northeast Pacific. Phycologia. 43(4):364–382.
- Heesch S, Broom JES, Neill KF, Farr TJ, Dalen JL, Nelson WA. 2009. Ulva, Umbraulva and Gemina: genetic survey of New Zealand taxa reveals diversity and introduced species. Eur J Phycol. 44(2):143–154.
- Hiraoka M, Ichihara K, Zhu WR, Shimada S, Oka N, Cui JJ, Tsubaki S, He PM. 2017. Examination of species delimitation of ambiguous DNA-based Ulva (Ulvophyceae, Chlorophyta) clades by culturing and hybridisation. Phycologia. 56(5):517–532.
- Kawai H, Shimada S, Hanyuda T, Suzuki T, Gamagori City Office. 2007. Species diversity and seasonal changes of dominant Ulva species (Ulvales, Ulvophyceae) in Mikawa Bay, Japan, deduced from ITS2 rDNA region sequences. Algae. 22(3):221–228.
- Kirkendale L, Saunders GW, Winberg P. 2013. A molecular survey of Ulva (Chlorophyta) in temperate Australia reveals enhanced levels of cosmopolitanism. J Phycol. 49(1):69–81.
- Kolmogorov M, Bickhart DM, Behsaz B, Gurevich A, Rayko M, Shin SB, Kuhn K, Yuan J, Polevikov E, Smith TPL, et al. 2020. MetaFlye: scalable long-read metagenome assembly using repeat graphs. Nat Methods. 17(11):1103–1110.
- Krupnik N, Rinkevich B, Paz G, Douek J, Lewinsohn E, Israel A, Carmel N, Mineur F, Maggs CA. 2018. Native, invasive and cryptogenic Ulva species from the Israeli Mediterranean Sea: risk and potential. Mediterranean Mar Sci. 19(1):132–146.
- Liu JL, Tong YC, Xia J, Sun YQ, Zhao XH, Sun JY, Zhao S, Zhuang MM, Zhang JH, He PM. 2022. Ulva macroalgae within local aquaculture ponds along the estuary of Dagu River, Jiaozhou Bay, Qingdao. Mar Pollut Bull. 174:113243.
- Loughnane CJ, McIvor LM, Rindi F, Stengel DB, Guiry MD. 2008. Morphology, rbcL phylogeny and distribution of distromatic Ulva (Ulvophyceae, Chlorophyta) in Ireland and Southern Britain. Phycologia. 47(4):416–429.
- National Standards of the People's Republic of China. 2007. Specifications for oceanographic survey. Part 6: marine biological survey. GB/T 12763.6.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Scagel RF, Gabrielson PW, Garbary DJ, Golden L, Hawkes MW, Lindstrom SC, Oliveira JC, Widdowson TB. 1989. A synopsis of the benthic marine algae of British Columbia, Southeast Alaska, Washington and Oregon. Canada: Phycological Contributions, University of British Columbia; p. 532.
- Sfriso A, Buosi A, Wolf MA, Sfriso AA. 2020. Invasion of alien macroalgae in the Venice Lagoon, a pest or a resource? Aquat Invasions. 15(2):245–270.
- Steinhagen S, Karez R, Weinberger F. 2019. Cryptic, alien and lost species: molecular diversity of Ulva sensu lato along the German coasts of the North and Baltic Seas. Eur J Phycol. 54(3):466–483.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Tanner CE. 1986. Investigations of the taxonomy and morphological variation of Ulva (Chlorophyta): Ulva californica Wille. Phycologia. 25(4):510–520.
- Verlaque M, Breton G. 2019. Biological invasion: long term monitoring of the macroalgal flora of a major European harbor complex. Mar Pollut Bull. 143:228–241.
- Wei X, Liu WZ, Lin XY, Liu QC, Jiang P. 2022. First record of Ulva californica in mainland China: a single alien parthenogenetic population with discontinuous distribution. J Oceanol Limnol.
- Wolf MA, Sciuto K, Andreoli C, Moro I. 2012. Ulva (Chlorophyta, Ulvales) biodiversity in the North Adriatic Sea (Mediterranean, Italy): cryptic species and new introductions. J Phycol. 48(6):1510–1521.
- Xie WF, Wu CH, Zhao J, Lin XY, Jiang P. 2020. New records of Ulva spp. (Ulvophyceae, Chlorophyta) in China, with special reference to an unusual morphology of U. meridionalis forming green tides. Eur J Phycol. 55(4):412–425.
