Abstract
Stephania epigaea H. S. Lo, 1978 is a medicinal plant commonly used in southwest China. This study characterized the first complete chloroplast (cp) genome sequence of this species. The complete cp was 157,738 bp in length, containing a large single-copy region (LSC) of 88,460 bp, a small single-copy region (SSC) of 19,778 bp, and a pair of inverted repeat regions (IRs) of 24,750 bp. It encoded 130 genes, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The GC content of the complete genome was 36.7%. Phylogenetic analysis of complete cp sequences revealed that S. epigaea was clustered with S. japonica from the Menispermaceae family.
Stephania epigaea H. S. Lo, 1978 (Menispermaceae) is distributed mainly in Yunnan and Sichuan provinces, China (Zhao et al. Citation2020; Dong et al. Citation2015), frequently used to treat cough, diarrhea, bellyache, injuries, and malaria (Xiao et al. Citation2021). Previous studies of this species mainly focused on its chemical constituents and medicinal properties (Lv et al. Citation2013; Dong et al. Citation2018). Two barcode analyses have been published using rDNA and a chloroplast marker (Xie et al. Citation2015; Wang et al. Citation2020). However, the complete chloroplast (cp) genome of this species has not yet been deciphered. Therefore, we reported the first complete cp genome of S. epigaea, which will provide a valuable resource for further genetic conservation, evolution and molecular breeding studies in the genus Stephania.
This article is licensed under the Regulations of Yunnan Province on Biodiversity Protection and is not require any ethical or institutional approvals. Healthy and fresh leaves of S. epigaea were collected from Dali county (25°84′ 95″ N, 100°11′6″ E) and deposited in the Herbarium of Dali University (http://yxy.dali.edu.cn/yhxy/, Min Fan, [email protected]) under the voucher number LJ2020060607. The total genomic DNA was extracted using the improved CTAB method (Doyle and Doyle Citation1986) and sequenced by the Illumina NovaSeq 6000 platform (Chang et al. Citation2021). In total, 19,098,862 clean reads (https://github.com/ndierckx/novoplasty) were de novo assembled by NOVOPlasty (Wang et al. Citation2020; Xia et al. Citation2021) and annotated by GeSeq with default settings (Tillich et al. Citation2017; Wei and Li Citation2021). The GenBank accession number is MZ678241.
The length of the S. epigaea cp genome was 157,738 bp with a typical quadripartite circular structure (Zhou et al. Citation2017), which contained a pair of inverted repeat regions (IRa and IRb, 24,750 bp), a large single-copy region (LSC, 88,460 bp), and a small single-copy region (SSC, 19,778 bp). All 130 genes were identified, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The overall GC content of the cp genome was 36.7%.
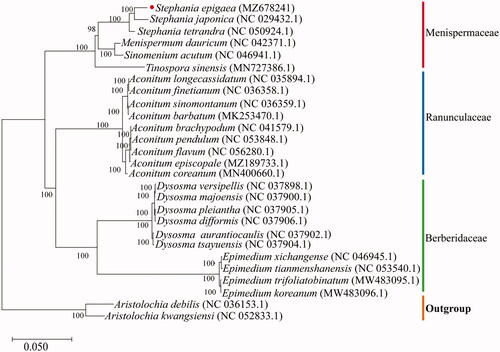
To evaluate the phylogenetic relationship of S. epigaea, 5 Menispermaceae, 10 Berberidaceae, and 9 Ranunculaceae complete cp genomes were downloaded from the NCBI database and aligned by MAFFT v7 (https://mafft.cbrc.jp/alignment/server/index.html) (Katoh and Standley Citation2013; Yamada et al. Citation2016). A maximum-likelihood tree with 1,000 bootstrap replicates was inferred using IQ-TREE v2.1.2 (Nguyen et al. Citation2015; Liu et al. Citation2021), with Aristolochia debilis (NC 036153.1) and Aristolochia kwangsiensis (NC 052833.1) as outgroups. The phylogenetic analysis revealed that S. epigaea was closely related to S. japonica (). The cp genome of S. epigaea will be helpful for a comprehensive understanding of phylogenetic relationships among the genus Stephania and provide a valuable reference for the conservation genetics of this species.
Author contributions statement
Qin Guan analyzed the data, prepared figures, authored drafts of the paper and approved the final draft. Danping Feng collected materials and performed the experiments. Min Fan designed the experiments, contributed reagents/materials/analysis tools, revised the content and approved the final draft. All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data, supporting the findings of this study, are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov) with the accession number is MZ678241, which permits unrestricted use, distribution and reproduction in any medium provided the original work is cited correctly. The associated BioProject, Bio-Sample and SRA numbers are PRJNA759697, SAMN21188258 and SRR15693952, respectively.
Additional information
Funding
References
- Chang QX, Li Y, Xia PG, Chen X. 2021. The complete chloroplast genome of Aster sampsonii (Hance) Hemsl, a perennial herb. Mitochondrial DNA B Resour. 6(9):2594–2595.
- Dong JW, Cai L, Fang YS, Xiao H, Li ZJ, Ding ZT. 2015. Proaporphine and aporphine alkaloids with acetylcholinesterase inhibitory activity from Stephania epigaea. Fitoterapia. 104:102–107.
- Dong JW, Li XJ, Cai L, Shi JY, Li YF, Yang C, Li ZJ. 2018. Simultaneous determination of alkaloids dicentrine and sinomenine in Stephania epigeae by 1H NMR spectroscopy. J Pharm Biomed Anal. 160(25):330–335.
- Doyle JJ, Doyle JI. 1986. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu S, Li YR, Si W, Qu WR, Yang TG, Wu ZH, Jiao PP. 2021. Complete chloroplast genome sequence of Oxytropis glabra (Leguminosae). Mitochondrial DNA B Resour. 6(9):2478–2479.
- Lv JJ, Xu M, Wang D, Zhu HT, Yang CR, Wang YF, Li Y, Zhang YJ. 2013. Cytotoxic bisbenzylisoquinoline alkaloids from Stephania epigaea. J Nat Prod. 76(5):926–932.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang J, Feng DP, Qian J, Duan BZ, Fan M. 2020. Characterization of the complete chloroplast genome of Salvia tiliifolia Vahl (Lamiaceae). Mitochondrial DNA B Resour. 5(3):2174–2175.
- Wang XL, Xue JY, Zhang YY, Xie H, Wang YQ, Weng WY, Kang Y, Huang JM. 2020. DNA barcodes for the identification of Stephania (Menispermaceae) species. Mol Biol Rep. 47(3):2197–2203.
- Wei YH, Li XP. 2021. Characterization of the complete chloroplast genome of Salix gordejevii (Salicaceae). Mitochondrial DNA B Resour. 6(9):2510–2512.
- Xia HH, Xu YF, Liao BR, Huang ZQ, Zhou HG, Wang FL. 2021. Complete chloroplast genome sequence of a Chinese traditional cultivar in Chrysanthemum, Chrysanthemum morifolium 'Anhuishiliuye. Mitochondrial DNA B Resour. 6(4):1281–1282.
- Xiao J, Wang YJ, Yang YQ, Liu JY, Chen G, Lin B, Hou Y, Li N. 2021. Natural potential neuroinflammatory inhibitors from Stephania epigaea H.S. Lo. Bioorg Chem. 107:104597.
- Xie DT, He JY, Huang JM, Xie H, Wang YQ, Kang Y, Jabbour F, Guo JX. 2015. Molecular phylogeny of Chinese Stephania (Menispermaceae) and reassessment of the subgeneric and sectional classifications. Aust Systematic Bot. 28(4):246.
- Yamada KD, Tomii K, Katoh K. 2016. Application of the MAFFT sequence alignment program to large data-reexamination of the usefulness of chained guide trees. Bioinformatics. 32(21):3246–3251.
- Zhao WL, Liu MY, Shen C, Liu HQ, Zhang ZT, Dai W, Liu XF, Liu JH. 2020. Differentiation, chemical profiles and quality evaluation of five medicinal Stephania species (Menispermaceae) through integrated DNA barcoding, HPLC-QTOF-MS/MS and UHPLC-DAD. Fitoterapia. 141:104453.
- Zhou JG, Chen XL, Cui YX, Sun W, Li YH, Wang Y, Song JY, Yao H. 2017. Molecular structure and phylogenetic analyses of complete chloroplast genomes of two Aristolochia medicinal species. IJMS. 18(9):1839.