Abstract
Rhododendron calophytum Franch is a famous ornamental plant, which belongs to the Rhododendron genus. Here, we report the complete chloroplast (cp) genome and the cp genomic features of R. calophytum. The complete cp genome of R. calophytum is a double stranded, circular DNA with 200,196 bp in length and has an large single-copy (LSC) region of 108,602 bp and a small single-copy (SSC) region of 2606 bp separated by a pair of inverted repeat (IR) regions of 44,494 bp each. Its 110 genes include four unique rRNAs, 29 tRNAs, and 77 protein-coding genes. Maximum-likelihood (ML) phylogenetic tree reconstructed with 16 species complete cp sequences reveals that R. calophytum is closely related to R. platypodum. The complete cp genome of R. calophytum will be useful for further investigations and researches concerning this economically important plant.
As a large plant branch, Rhododendron is the largest genus in Ericaceae, comprising 1025 species of woody plants, either evergreen or deciduous (Chamberlain et al. Citation1996). Rhododendron species are widely distributed throughout America, Europe, since their flowers are attractive and bright, many Rhododendron species are valued as ornamental plants in landscaping (Yan et al. Citation2015). Rhododendron calophytum Franch. Bull. Soc. Bot. France (1886) is an important economic tree species because it contains a variety of flavonoids and volatile oil active components with high and medicinal value (Tian et al. Citation2010). Lacking of molecular markers and genome information limits R. calophytum development and utilization. In this study, we presented the complete chloroplast (cp) genome sequence of R. calophytum based on Illumina sequencing and the phylogenetic relationship with Ericaceae relative species.
Sample of R. calophytum was collected from the Wolong National Nature Reserve in Sichuan Province (E: 102.97427°, N: 30.85641°, and H: 2826 m). Vouchers were deposited at West China Subalpine Botanical Garden, Dujiangyan Sichuan Province (2018-WL-009). We isolated the genomic DNA from silica gel-dried leaves of an individual of R. calophytum, according to the modified CTAB method (Doyle Citation1987). Purified genomic DNA was sequenced using an Illumina HiSeq X Ten sequencer. Overall, 6G clean reads were obtained after Illumina sequencing and trimmed by NGSQC (Patel and Jain Citation2012) with default parameters. We used MITObim v1.8 (Hahn et al. Citation2013) and Novoplasty v2.6.7 (Dierckxsens et al. Citation2017) to assemble with the cp genomes by comparing with the cp genomes of R. pulchrum (MN182619.1) (Shen et al. Citation2019) and R. platypodum (NC_053746.1) (Ma et al. Citation2021). After manual adjustment, a 200,196 bp cp genome was obtained and an automated genome annotation was performed using DOGMA (Wyman et al. Citation2004). The annotated cp genome of R. calophytum has been deposited in GenBank with the accession number OM373082. The complete cp genome of R. calophytum is a double stranded, circular DNA 200,196 bp in length and has an large single-copy (LSC) region of 108,602 bp and a small single-copy (SSC) region of 2606 bp separated by a pair of inverted repeat (IR) regions of 44,494 bp each. Its 110 genes include four unique rRNAs, 29 tRNAs, and 77 protein-coding genes. Most cp genes are single copy, while 39 genes are double copies, including four rRNA (4.5S, 5S, 16S, and 23S rRNA), nine tRNA, and 16 PCGs. The gene trnI-CAU has four copies in cp genome. The overall GC content of cp genome of R. calophytum is 35.9%.
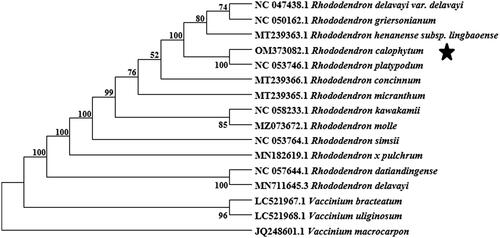
To confirm R. calophytum phylogenetic position in the Rhododendron, there were 16 cp genomes of 13 Rhododendron species, and three outgroups species Vaccinium bracteatum, V. macrocarpon, and V. uliginosum. The sequences were aligned using MAFFT v7.471 (Katoh et al. Citation2002). Maximum-likelihood (ML) phylogenetic tree was reconstructed with MEGA X (Kumar et al. Citation2018) with 1000 bootstrap replicates and the GTR + G model. The phylogenetic analysis reveals that all sampled species of Rhododendron clustered into a monophyletic clade with high bootstrap supporting values. The R. calophytum is closely related to R. platypodum (). This complete cp genome can be subsequently used for population genomic studies of R. calophytum and phylogenetic reconstruction of Ericaceae and will be helpful for future studies regarding the evolution of related taxa.
Figure 1. Phylogeny of 15 species within the order Ericaceae based on the maximum-likelihood (ML) analysis of complete chloroplast genome. The bootstrap values were based on 1000 replicates, and are indicated next to the branches. The taxa included as outgroups are Vaccinium bracteatum, V. macrocarpon, and V. uliginosum.
Ethics statement
The authors declare that all experiments described herein comply with the law of government in which they were carried out. All plant materials used in this study are legally obtained and/or collected.
Author contributions
Wenbao Ma: methodology, data curation, formal analysis, software, and writing-manuscript. Tao Yu: methodology and writing-manuscript. Wanjiang Wang: methodology, Fei Wang: methodology, Changlong Mu: investigation, methodology, and funding acquisition. All authors agreed to the published version of the manuscript.
Disclosure statement
The authors report no conflict of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OM373082. The accession numbers for these SRA data in GenBank of NCBI are as follows: SRA: SRR17867967, BioProject: PRJNA802982, and BioSamples: SAMN25602456.
Additional information
Funding
References
- Chamberlain D, Hyam R, Argent G, Fairweather G, Walter KS. 1996. The genus Rhododendron: its classification and synonymy. Edinburgh: Royal Botanic Garden.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Ma LH, Zhu HX, Wang CY, Li MY, Wang HY. 2021. The complete chloroplast genome of Rhododendron platypodum (Ericaceae): an endemic and endangered species from China. Mitochondrial DNA B Resour. 6(1):196–197.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619.
- Shen JS, Li XQ, Zhu XT, Huang XL, Jin SH. 2019. Complete chloroplast genome of Rhododendron pulchrum, an ornamental medicinal and food tree. Mitochondrial DNA B Resour. 4(2):3527–3528.
- Tian P, Fu X, Zhuang P, Bai J, Chen F. 2010. Analysis on volatile oil from Rhododendron calophytum Franch by GC–MS. Chin J Appl Environ Biol. 16(5):734–737.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yan LJ, Liu J, Moller M, Zhang L, Zhang XM, Li DZ, Gao LM. 2015. DNA barcoding of Rhododendron (Ericaceae), the largest Chinese plant genus in biodiversity hotspots of the Himalaya–Hengduan Mountains. Mol Ecol Resour. 15(4):932–944.
