Abstract
Phalaenopsis malipoensis Z.J. Liu et S.C. Chen, 2005 is a valuable species of Orchidaceae with significant ornamental value. This study reports the first complete chloroplast genome of P.malipoensis was collected integrally and clarified further. The genome was 144,528 bp in length and comprised four regions, including a pair of inverted repeats each 24,710 bp, a large single-copy region of 83,475 bp, and a small single-copy region of 11,633 bp. The whole genome contained 125 genes, including 8 rRNAs, and 39 tRNAs. Phylogenetic analysis indicated that P. malipoensis was closely related to Phalaenopsis lobbii. The analysis of P. malipoensis chloroplast genome will provide significant data for the identification and further improvement of P. malipoensis.
Orchidaceae is the largest family of monocotyledons plant family, with abundant floral resources, including five subfamilies (Chase et al. Citation2015). The genus Phalaenopsis has butterfly-like flowers, bright colors, and a long flowering period. It has the most popular and highly economical orchid species (Handini et al. Citation2016). It has a long history of cultivation, through years of breeding, has been cultivated with many good characters, and high ornamental values varieties (Antonetti et al. Citation2021). However, with the destruction of the native habitats and reckless human pilfering, the number of native populations has been constantly decreasing. The Phalaenopsis malipoensis Z.J. Liu et S.C. Chen, 2005 is a new species of Phalaenopsis found in Malipo County, Yunnan Province, China, 2005 (Liu et al. Citation2005). It has a three-lobed lip, both white and yellow, and is very similar to Phalaenopsis lobbii in flower shape and color. (Liu et al. Citation2005; Zhang et al. Citation2020). This precious and rare species was endemic to China, but the lack of detailed information and necessary conservation measures has hindered its exposure to the world. Therefore, we studied the entire chloroplast genome of P. malipoensis and mapped its genetic and genomic data for use in genetics, phylogeny, and breeding of the Phalaenopsis.
Samples of P. malipoensis for sequencing were collected from Yunnan Province, China, and the living plants were deposited at the National Orchid Conservation Center of Fujian Agriculture and Forestry University (26°20′21.3″N, 113°12′39.6″E, Voucher specimen: MLPHDL-YN2019-19A, Yuzhen Zhou, [email protected]). DNA was extracted from leaves by hexadecyltrimethylammonium bromide (CTAB) method. The clean reads were obtained by sequencing through the Illumina NovaSeq high-throughput sequencing platform. The sequencing generated about 16 GB of data, which was stored in the GenBank Sequence Read Archive (SRA) (accession number: SRR17270921). Low quality reads and adapters were removed using the Fastp v0.23.1 software with default parameters (Chen et al. Citation2018). Sequence assembly was performed using Get Organelle (core software SPAdes 3.1; assembly options: -R 15 -k 21, 45, 65, 85, 105) (Bankevich et al. Citation2012). The manual correction was done using Bandage v0.8.1 software, as previously reported (Wick et al. Citation2015). The refined genome was annotated by using the online facilities, CPGAVAS2 and Geneious Prime v2022.0.1 programming (reference: Phalaenopsis lobbii, NC_059699.1), followed by physical check for the corrections of explanation data. Complete chloroplast sequence of P. malipoensis (GenBank accession: OL623704) is 144,528 bp in length with an average GC content of 37%. It contains a large single-copy (LSC) region of 83,475 bp, a small single-copy (SSC) region of 11,633 bp and a pair of inverted repeat (IR) of 24,710 bp. It contains a total of 125 genes, including 8 rRNAs and 38 tRNAs.
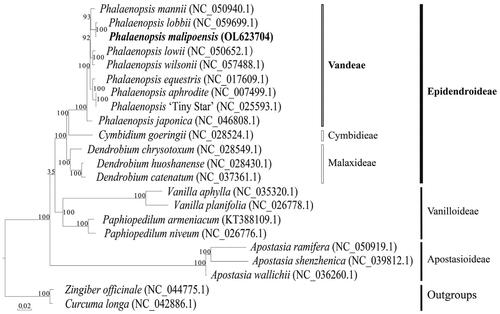
To determine the location of the phylogenetic tree, the sequences of P. malipoensis were compared with other 21 chloroplast genomic sequences downloaded from the NCBI GenBank using MAFFT v7 (Katoh et al. Citation2019). Comparison of the P. malipoensis chloroplast genome to previously published data shows a high level of gene synteny with one publicly available genome of P. Lobbii (Zhang et al. Citation2020). Maximum likelihood tree was constructed with 1000 bootstrap replicates through the RAxML-HPC2 on XSEDE program available on the CIPRES online website (https://www.phylo.org/). The output files were visualized by EvolView v3 with Zingiber officinale (NC_044775.1) and Curcuma longa (NC_042886.1) as outgroups (Alexandros Citation2014; Balakrishnan et al. Citation2019). The result of the phylogenetic tree indicated that P. malipoensis, P. Lobbii, and Phalaenopsis mannii (NC_050940.1) were sister groups () (Chen et al. Citation2020). Thus, the chloroplast genome analysis of P. malipoensis provides supportive data to further research on conservation genetics, phylogeny, and molecular breeding of Orchids.
Figure 1. Maximum-likelihood phylogenetic tree include the complete chloroplasts of Phalaenopsis malipoensis (OL623704) and 19 other orchids with 2 outgroups, Zingiber officinale (NC_044775.1) and Curcuma longa (NC_042886.1). P. malipoensis is indicated in bold italic. The number on the branch node represents the bootstrap percentage of maximum parsimony, and the GeneBank accession number is shown after the scientific name of the species. Subfamilies of Orchidaceae are shown on the right.
Ethical approval
The material covered in the article, all collection and sequencing were carried out in strict accordance with local laws and relevant laboratory regulations to protect wild resources. This study was permitted by the Key Laboratory of National Forestry and Grassland Administration for Orchid Conservation and Utilization, FAFU, China.
Authors’ contributions
YZZ. and KZ managed the study; YHC and MJH planned and coordinated the study; YPZ and YT collected plant samples for sequencing; YTZ, WJW, EHT and MJH participated in data analysis; YHC, YPZ and MJH drafted the manuscript; YHC, YZZ and KZ revised the manuscript. All authors have provided commentary and final approval.
Collection manager email
A specimen was deposited at the National Orchid Conservation Center of Fujian Agriculture and Forestry University (Yuzhen Zhou, [email protected]) under the voucher number MLPHDL-YN2019-19A.
Acknowledgment
We are grateful to The National Orchid Conservation Centre of China, Yang Hao, Dingkun Liu and Dr. Sagheer Ahmad for their advice and assistance with this project.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/WebSub/) under the accession no. OL623704. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA790458, SRR17270921, and SAMN24199872 respectively.
Additional information
Funding
References
- Alexandros S. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Antonetti M, Nin S, Burchi G, Biricolti S, Gori M. 2021. Himantoglossum adriaticum H. Baumann × Himantoglossum robertianum (Loisel.) P. Delforge: A New Interspecific Hybrid Assessed by Barcoding Analysis. Plants. 10(1):107–125.
- Balakrishnan S, Gao S, Lercher MJ, Hu S, Chen WH. 2019. Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nuclc Acids Research. 47:270–275.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology. 19(5):455–477.
- Chase MW, Cameron KM, Freudenstein JV, Pridgeon AM, Salazar G, van den Berg C, Schuiteman A. 2015. An updated classification of Orchidaceae. Bot J Linn Soc. 177(2):151–174.
- Chen B, Zhang Y, Cao Y, Zheng Y, Wei Z, Zhao K, Peng D, Zhou Y. 2020. Chloroplast characterizations of a Phalaenopsis native to China, Phalaenopsis mannii (Orchidaceae). Mitochondrial DNA Part B. 5(3):3707–3708.
- Chen S, Zhou Y, Chen Y, Jia G. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–890.
- Handini AS, Sukma D, Sudarsono D. 2016. Analisis Keragaman Morfologi dan Biokimia pada Anggrek Phalaenopsis (Orchidaceae). J Agron Indonesia. 44(1):62–67.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Liu ZJ, Chen SC, Zheng-Zhong RU. 2005. Phalaenopsis malipoensis, a New Species of Orchidaceae from China. Acta Botanica Yunnanica. 27:37–38.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Zhang Y, Chen B, Zheng Y, Cao Y, Wei Z, Zhao K, Zhou Y. 2020. Characterization of the complete chloroplast genome of Phalaenopsis lobbii (Orchidaceae), an important horticultural plant in China. Mitochondrial DNA Part B. 5(3):3468–3469.
