Abstract
We determined the complete mitochondrial genome sequence of a holothurian Euapta godeffroyi belonging to the order Apodida. The complete mitogenome of E. godeffroyi was 16,410 bp in length and consisted of 13 protein-coding genes (PCGs), two ribosomal RNA genes, and 22 transfer RNA genes. The orders of PCGs and rRNAs did not match those of any recorded holothurian mitogenomes. The maximum likelihood (ML) phylogenetic tree placed E. godeffroyi as the sister group to chiridotid species and supported the monophyly of the order Apodida.
Euapta godeffroyi (Semper, Citation1868) () is a common sea cucumber in coral reefs in the tropical Indo-Pacific region and shows geographically large distribution in the tropical shallow waters from the west Indian Ocean to the east Pacific Ocean (e.g., Massin Citation1999). Apodid holothurians including E. godeffroyi are a stem group within the class Holothuroidea (Kerr and Kim Citation2001; Miller et al. Citation2017). Miller et al. (Citation2017) confirmed the monophyly of the order Apodida, and that apodid holothurians are divided into two clades in the order Apodida: (1) family Myriotrochidae and (2) other apodid holothurians. In the later clade, however, the family Synaptidae including E. godeffroyi did not form a monophyletic clade. A part of synaptid species, Leptosynapta clarki Heding, 1928, was placed in Chiridotidae. Their molecular phylogenetic study of Miller et al. (Citation2017) used six mitochondrial/nuclear gene markers, covering all common gene markers for Echinodermata, but their study included limited OTUs from apodid holothurians, and the relationships between L. clarki and chiridotid species lacked strong nodal support. Therefore, the order Apodida requires the phylogenetic and systematic revision using further genetic markers and OTUs, and 13 protein-coding genes of mitogenome are expected to be effective in revising the phylogenetic relationship of apodid holothurians. There has been no mitogenome record for the family Synaptidae in Apodida, and we choose E. godeffroyi as representative of the family. We determined the whole mitogenome sequence by a shotgun sequencing method.
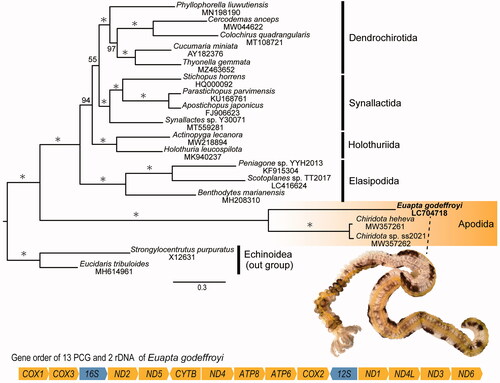
Figure 1. Molecular phylogenetic tree based on holothurian mitogenomes including Euapta godeffroyi, and gene order of 13 protein-coding genes (PCG) and two ribosomal DNA (rDNA) in the mitogenome of E. godeffroyi. Maximum-likelihood tree was analyzed by the concatenated nucleotide sequence of 13 PCG of E. godeffroyi (noted in bold type, LC704718) and 16 holothuroid and two echinoid species. Values on branches are ML bootstrap support values (BS) and asterisks (*) indicate 100%. The scale bar indicates branch length in substitutions per site. The accession numbers and references for all species are as follows: Actinopyga lecanora, MW218894#; Apostichopus japonicus, FJ906623, Sun et al. Citation2010; Benthodytes marianesis, MH208310, Mu et al. Citation2018; Cercodemas anceps, MW044622, Li et al. 2021; Chiridota heheva, MW357261, Sun et al. Citation2021; Chiridota sp. SS-2021, MW357262, Sun et al. Citation2021; Colochirus quadrangularis, MT108721, Zeng et al. Citation2020; Cucumaria minuta, AY182376, Scouras et al. Citation2004; Holothuria leucospilata, MK940237, Yang et al. Citation2019; Parastichopus parvimensis, KU168761#; Peniagone sp. YYH-2013, KF915304#; Phyllophorella liuwutiensis, MN198190, Yang et al. Citation2020; Scotoplanes sp., LC416624, Takano et al. Citation2019; Stichopus horrens, HQ000092, Fan et al. Citation2011; Synallactes sp. Y30071, MT559281, Liao et al. Citation2020; Thyonella gemmata, MZ463652, Figueroa et al. Citation2021; Strongylocentrutus purpuratus, X12631, Jacobs et al. Citation1988; and Eucidaris tribuloides, MH614961#. Octothorpes (#) mean direct submitted sequences in GenBank.
The specimen was collected in 2019 from North Point, Nyaung Oo Phee Island, Myanmar, facing the Andaman Sea (10.087 N 97.963 E), at the depth of approximately 22 m. Total DNA was extracted using DNeasy Blood & Tissue Kit (QIAGEN) and processed using the Collibri™ PS DNA Library Prep Kits for Illumina Systems (Invitrogen). Paired-end sequencing (300 cycles) was conducted using HiSeqX (Illumina) of Macrogen Japan Corp., with 150 bp read length and additional inserts of ca. 100 bp for a total of approximate 16 million reads. Assembly was performed using CLC Genomics Workbench ver. 12 (QIAGEN) with the default setting. Gene identification was made using the MITOS web server (Bernt et al. Citation2013). The voucher specimen is deposited in the National Museum of Nature and Science, Tsukuba, Japan (NSMT E-13912).
The mitogenome of E. godeffroyi (GenBank/DDBJ/EMBL accession number LC704718) is 16,410 bp long and encodes 13 proteins, two rRNAs, and 22 tRNAs for a total of 37 gene products. The overall A + T content is 64.0%, which is slightly lower than that of other apodid species (Sun et al. Citation2021). Unlike other sea cucumber mitogenomes, COX1, ND4L, and ND1 start with GTG codon. All other protein-coding genes (PCGs) start with the ATG start codon. Eight of PCGs stop with the termination codon TAG, other five of PCGs (CYTB, ND2, ND3, ND4, and ATP6) end with TAA codon. Although previously three variations of the gene orders of 13 PCGs and rRNAs were known in the class Holothuroidea (Sun et al. Citation2021), the gene order of E. godeffroyi did not match those of any recorded holothurian mitogenomes. The gene order of E. godeffroyi shows unique arrangement with COX3 following after COX1, and ND3 located next to ND6 ().
The maximum-likelihood phylogenetic analysis (ML) based on 13 PCGs of E. godeffroyi and 16 other holothurians (Scouras et al. Citation2004; Sun et al. Citation2010, Citation2021; Fan et al. Citation2011; Mu et al. Citation2018; Takano et al. Citation2019; Yang et al. Citation2019, Citation2020; Liao et al. Citation2020; Zeng et al. Citation2020; CitationFigueroa et al. 2021; Li et al. Citation2021; three other direct submitted sequences in GenBank), and of two echinoids (Jacobs et al. Citation1988; another direct submitted sequence in GenBank) as outgroup, was conducted using RAxML-NG ver.1.0.2 (Kozlov et al. Citation2019) with bootstrap analyses of 1000 replicates (). PartitionFinder 2.1.1 (Lanfear et al. Citation2017) was used to determine the best partitioning scheme and the substitution model with branch lengths linked and a greedy search algorithm (Lanfear et al. Citation2012). The optimal partitioning strategy and evolutionary models consisted of thirteen genes data set for ML analyses were as follows; partition 1 (COX1), partition 2 (COX2), partition 3 (COX3), partition 4 (CYTB), partition 5 (ND1), partition 6 (ND2 and ND4), partition 7 (ND3, ND4L, and ATP6), partition 8 (ND5), and partition 9 (ATP8) with GTR + I + G; partition 9 (ND6) with K81UF + I + G. Three apodids, E. godeffroyi, Chiridota heheva and Chiridota sp., form a monophyletic clade, and E. godeffroyi is a sister taxon to the other Apodida (C. heheva and Chiridota sp.) with high nodal support (). This relationship between the three apodid species in our tree was consistent with the result of Miller et al. (Citation2017). Although additional OTUs are needed, this mitogenome will be useful for reconstructing higher systematics of Holothuroidea phylogeny.
Ethics statement
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Author’s contributions
All authors conceived the study. MMA and TF contributed to the application for sampling permits and conducted the voucher specimen collection and the DNA sample preparation in Myanmar. AO and SFH performed DNA analysis in the laboratory. All authors were involved in the interpretation of the data; the drafting of the paper, revising it critically for intellectual content; and the final approval of the version to be published; and that all authors agree to be accountable for all aspects of the work.
Acknowledgments
We are grateful to Forest Department, Ministry of Natural Resources and Environmental Conservation for their permission to carry out field works in reserved forests, and for their support and collaboration.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The consensus genome sequence is openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. LC704718. The associated BioProject, SRA, and Bio-Sample numbers are PRJDB13488, DRR361674, and SAMD00467977 respectively. The voucher specimen is deposited to the National Museum of Nature and Science, Tsukuba, Japan (Toshihiko Fujita; [email protected]) under the catalog number NSMT E-13912.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Fan S, Hu C, Wen J, Zhang L. 2011. Characterization of mitochondrial genome of sea cucumber Stichopus horrens: a novel gene arrangement in Holothuroidea. Sci China Life Sci. 54(5):434–441.
- Figueroa AC, McHugh WJ, Miller SM, Fellgren AK, Bogantes VE, Janosik AM. 2021. Characterization of the complete mitochondrial genome of Thyonella gemmata (Echinodermata: Cucumariidae). Mitochondrial DNA B Resour. 6(10):2997–2998.
- Jacobs HT, Elliott DJ, Math VB, Farquharson A. 1988. Nucleotide sequence and gene organization of sea urchin mitochondrial DNA. J Mol Biol. 202(2):185–217.
- Kerr AM, Kim J. 2001. Phylogeny of Holothuroidea (Echinodermata) inferred from morphology. Zoological Journal of the Linnean Society. 133(1):63–81. doi:10.1111/j.1096-3642.2001.tb00623.x.
- Kozlov A, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455.
- Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Li H, Liu J, Wang S, Huang W. 2021. The complete mitochondrial genome of Pink warty sea cucumber (Cercodemas anceps Selenka, 1867). Mitochondrial DNA B Resour. 6(3):959–961. doi:10.1080/23802359.2021.1891979.
- Liao M, Li B, Xiao N, Kong M, Wang Y, Wang J, Rong X, Zhang Z, Yu Y. 2020. Complete sequence of mitochondrial DNA of a deep-sea holothurian species of the genus Synallactes (Synallactida: Synallactidae). Mitochondrial DNA B Resour. 5(3):2699–2700. doi:10.1080/23802359.2020.1787266.
- Massin C. 1999. Reef-dwelling Holothuroidea (Echinodermata) of the Spermonde Archipelago (South-West Sulawesi, Indonesia). Zoo Verh. 329:1–144.
- Miller AK, Kerr AM, Paulay G, Reich M, Wilson NG, Carvajal JI, Rouse GW. 2017. Molecular phylogeny of extant Holothuroidea (Echinodermata). Mol Phylogenet Evol. 111:110–131.
- Mu W, Liu J, Zhang H. 2018. Complete mitochondrial genome of Benthodytes marianensis (Holothuroidea: Elasipodida: Psychropotidae): insight into deep sea adaptation in the sea cucumber. PLoS ONE. 13(11):e0208051.
- Scouras A, Beckenbach K, Arndt A, Smith MJ. 2004. Complete mitochondrial genome DNA sequence for two ophiuroids and a holothuroid: the utility of protein gene sequence and gene maps in the analyses of deep deuterostome phylogeny. Mol Phylogenet Evol. 31(1):50–65.
- Semper C. 1868. Holothurien. In: von Engelmann, W. Reisen im Archipel der Philippinen. Zweiter Theil. Wissenschaftliche Resultate vol. 1., Leipzig, pp. 288.
- Sun S, Sha Z, Xiao N. 2021. The first two complete mitogenomes of the order Apodida from deep-sea chemoautotrophic environments: new insights into the gene rearrangement, origin and evolution of the deep-sea sea cucumbers. Comp Biochem Physiol D Genom Proteom. 39:100839.
- Sun XJ, Li Q, Kong LF. 2010. Comparative mitochondrial genomics within sea cucumber (Apostichopus japonicus): provide new insights into relationships among color variants. Aquaculture. 309(1-4):280–285.
- Takano T, Ijichi M, Itoh H, Fukuda H, Yoshizawa S. 2019. Complete mitochondrial genome sequences of a deep-sea holothurian species of the genus Scotoplanes (Elasipodida: Elpidiidae). Mitochondrial DNA B: Resour. 4(1):112–113.
- Yang F, Zhou C, Tran NT, Sun Z, Wu J, Ge H, Lu Z, Zhong C, Zhu Z, Yang Q, et al. 2020. Comparison of the complete mitochondrial genome of Phyllophorus liuwutiensis (Echinodermata: Holothuroidea: Phyllophoridae) to that of other sea cucumbers. FEBS Open Bio. 10(8):1587–1600.
- Yang Q, Lin Q, Wu J, Tran NT, Huang R, Sun Z, Zhu Z, Lu Z, Li S, Zhou C. 2019. Complete mitochondrial genome of Holothuria leucospilata (Holothuroidea, Holothuriidae) and phylogenetic analysis. Mitochondrial DNA B Resour. 4(2):2751–2752.
- Zeng L, Wen J, Lin H, Fan S, Sun Y, Yang C, Zhao J, Li X. 2020. The complete mitochondrial genome of Colochirus quadrangularis (Dendrochirotida, Cucumariidae). Mitochondrial DNA B: Resour. 5(2):1665–1666.
