Abstract
Verbascum thapsus L. has extensive pharmacological effects, including antioxidative and antineoplastic action, memory improvement and neuroprotection. However, its phylogenetic position is not established in Scrophulariaceae. In this study, we reported the complete chloroplast genome sequence of V. thapsus L. for the first time and investigate its phylogenetic relationship in Scrophulariaceae. The assembled chloroplast genome is a circular 153,338 bp sequence, including a large single copy (LSC) region of 84,627 bp, a small single copy (SSC) region of 17,829 bp and a pair of inverted repeats (IRs) of 25,441 bp. The genome contains 135 genes, including 86 protein coding genes, 37 tRNA genes, and eight rRNA genes. The phylogenetic tree showed that V. thapsus is closely associated with V. chinense and V. phoeniceum.
The genus Verbascum (Scrophulariaceae) contains about 300 species with medicinal properties, most of which are distributed across Europe and Asia. Currently, there are six Verbascum species recognized in China (Süntar et al. Citation2011; Ntantiso and Jaca Citation2015; Frezza et al. Citation2019). Among these species, Verbascum thapsus Linnaeus 1753 is widely distributed in southwest China and has been used in ethnic medicine for treatment of osteoporosis and depression (Minru and Yi Citation2016; Frezza et al. Citation2019). Until now, however, most studies on V. thapsus have concentrated on morphology and phytochemistry rather than molecular evolutionary history (Babamoradi et al. 2018). Chloroplast (cp) genomes are precious resources for phylogenetic studies and plant molecular identification (Wei et al. Citation2020). With such data, the phylogenetic positions of two Verbascum species have been clarified (Bi et al. 2020; He et al. Citation2020). In this study, we report the complete chloroplast genome sequence of V. thapsus and elucidated its phylogenetic position in Scrophulariaceae. This provides the basis for further study on the genetic diversity, phylogenetic relationships, and evolution of V. thapsus.
Fresh and young leaves of V. thapsus were collected from Lijiang, Yunnan, China (N26°55′33.88″, E100°14′09.02″). A voucher specimen was deposited in the Culture and Tourism College of Yunnan University (www.lywhxy.com, Contact person: Ai-en Tao, and Email: [email protected]) under the voucher number: TC01. Total genomic DNA was extracted from fresh leaves using a modified CTAB method (Doyle and Doyle Citation1987; Yang et al. Citation2014) and genome sequencing of 150 bp paired-end reads was performed using the Illumina Hiseq 2500 platform by Genesky Biotechnologies Inc. (Shanghai, China). About 4 G data were obtained and assembled with GetOrganelle v1.6.2 (Jin et al. Citation2020), and annotated with the OGAP pipeline (https://github.com/zhangrengang/ OGAP). The draft annotations were then adjusted manually. The annotated genomic sequence has been submitted to GenBank (accession number: MT012419).
The complete chloroplast genome of V. thapsus is 153,338 bp in length with a typical quadripartite structure, which contained a pair of inverted repeat regions (IRa and IRb) of 25,441 bp, and a large single copy (LSC) region of 84,627 bp, a small single copy (SSC) region of 17,829 bp. The content of guanine(G) and cytosine(C) in the whole chloroplast genome is 36.0%. In addition, The cp genome contained 135 genes, including 86 protein coding genes, 37 tRNA genes, and eight rRNA genes.
To investigate its phylogenetic position, we used complete chloroplast genome sequences from V. thapsus and 27 other Scrophulariaceae species downloaded from GenBank. The resulting phylogenetic tree was rooted with two Salvia species. These chloroplast genome sequences were aligned using MAFFT v.7.471 (Katoh and Standley Citation2013). Phylogenetic analysis was conducted with maximum likelihood and 1000 bootstrap replicates using IQTree (Nguyen et al. Citation2015). Bootstrap values were calculated using the in-built UFBoot within IQTree, and the best nucleotide substitution model, GTR + R6, was also estimated in IQTree. The phylogenetic reconstruction revealed V. thapsus formed a well-supported clade with two other Verbascum species (). This relationship will require further phylogenetic studies with additional taxa from Verbascum and related genera.
Figure 1. Maximum-likelihood (ML) phylogenetic tree for V. thapsus based on 30 complete chloroplast genomes. The numbers on the nodes indicate bootstrap values from 1000 replicates.
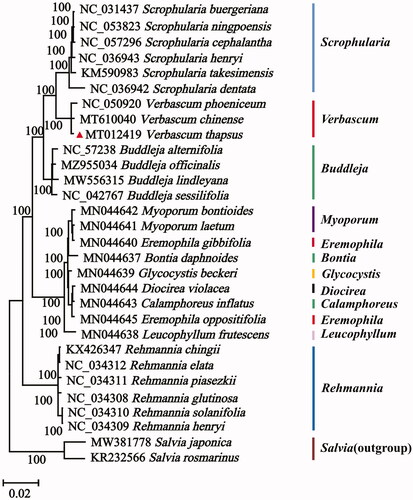
In this study, we sequenced and annotated the cp genome of V. thapsus and used these data in a phylogenetic analysis. This provides a valuable genomic resource for future research on the evolutionary history and conservation of Verbascum species.
Ethical approval
This article is licensed under a Regulations of Yunnan Province on biodiversity protection and approved by Lijiang counties(Yunnan Province, China), Dali University(Yunnan province of China).
Author contributions statement
Yue Zhang performed the experiments, analyzed the data, prepared figures and tables, authored drafts of the paper, and approved the final draft. Yun-hui Guan analyzed the data, prepared figures and tables. Cheng-zhao Pu, Yi-jia Xi analyzed the data. Cong-long Xia contributed reagents/materials/analysis tools, Ai-en Tao approved and approved the final draft.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, under the accession no. MT012419. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA798180, SRR17653644, SAMN25049704, respectively.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Frezza C, Bianco A, Serafini M, Foddai S, Salustri M, Reverberi M, Gelardi L, Bonina A, Bonina FP. 2019. HPLC and NMR analysis of the phenyl-ethanoid glycosides pattern of Verbascum thapsus L. cultivated in the Etnean area. Nat Prod Res. 33(9):1310–1316.
- He Y, Ma Y, Li Z, Liu P, Yuan W. 2020. The complete chloroplast genome of Verbascum chinense (L.) Santapau. Mitochondrial DNA Part B. 5(3):3039–3040.
- Süntar I, Tatlı II, Küpeli Akkol E, Keleş H, Kahraman Ç, Akdemir Z. 2011. An ethnopharmacological study on Verbascum species: from conventional wound healing use to scientific verification. J Ethnopharmacol. 132(2):408–413.
- Minru J, Yi Z. 2016. Dictionary of Chinese ethnic medicine. Beijing: China Medical Science and Technology Press.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Ntantiso Z, Jaca TP. 2015. Assessment of potential invasiveness of Verbascum thapsus L. in South Africa. South Afr J Bot. 98:214–215.
- Wei F, Tang DF, Wei KH, Qin F, Li LX, Lin Y, Zhu YX, Khan A, Kashif MH, Miao JH. 2020. The complete chloroplast genome sequence of the medicinal plant Sophora tonkinensis. Sci Rep. 10(1):12473.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14(5):1024–1031.
