Abstract
Hevea pauciflora belongs to the Euphorbiaceae family, an important wild relative of the rubber tree. This study sequenced, assembled, and annotated the complete chloroplast genome of H. pauciflora. The complete chloroplast genome is 161,123 bp with a canonical quadripartite structure containing a large single-copy (LSC) region (89,109 bp), a small single-copy (SSC) region (18,376 bp), and two inverted repeat regions (IRa and IRb) (26,819 bp, each). A total of 134 genes were annotated, including 86 protein-coding genes, four pseudogenes, 36 tRNA genes, and eight rRNA genes. The 134 genes include four major groups: ‘self-replication’, ‘photosynthesis’, ‘unknown function’, and ‘others’. A phylogenetic analysis clustered H. pauciflora, H. brasiliensis, H. camargoana, and H. benthamiana into one clade, consistent with traditional taxonomy. This study provides useful data for further studies of Hevea genus and the phylogenetic relationships of Euphorbiaceae species.
Hevea pauciflora (Benth.) Mull. Arg., Linnaea 1865, belongs to the Hevea genus of the Euphorbiaceae family, an important wild relative of the rubber tree (Hevea brasiliensis). The rubber tree is the primary commercial source of high-quality natural rubber, an important industrial raw material. The rubber tree and H. pauciflora originated from the Amazon river basin of South America. Nonetheless, the current rubber tree planting area mainly covers Indonesia, Malaysia, Thailand, China, and other Southeast Asian countries. Genus Hevea consists of 11 species (Clément-Demange et al. Citation2007), including H. pauciflora, H. brasiliensis, H. guianensis, H. camargoana, H. benthamiana, H. microphylla, H. rigidifolia, H. spruceana, H. paludosa, H. nitida, and H. camporum (Goncalves et al. Citation1990; Priyadarshan and Goncalves Citation2002). All the 11 species are diploids with 36 chromosomes, except H. pauciflora and H. guianensis, with 18 and 54 chromosomes, respectively (Baldwin Citation1947; Majumder Citation1964).
The nuclear genome of H. brasiliensis has been completely assembled (Rahman et al. Citation2013; Lau et al. Citation2016; Tang et al. Citation2016; Pootakham et al. Citation2017; Liu et al. Citation2020). Meanwhile, the chloroplast genomes of H. brasiliensis (Tangphatsornruang et al. Citation2011), H. benthamiana (Niu et al. Citation2020), and H. camargoana (Niu et al. Citation2020) have been sequenced. H. pauciflora, the only species with 18 chromosomes in the genus Hevea, has great potential for distant hybridization breeding of the rubber tree. However, the chloroplast genome sequence of H. pauciflora, a crucial requirement for hybridization breeding, was lacking. Thus, this study sequenced and analyzed the chloroplast genome of H. pauciflora.
Healthy, young leaves of H. pauciflora () were collected from The Rubber Tree Germplasm Resource Nursery of the Chinese Academy of Tropical Agriculture Science (N 19°34′31.53″ and E 109°31′17.97″). High-quality genomic DNA was extracted from H. pauciflora leaves using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. The specimens and DNA samples were deposited in the herbarium and cryogenic sample library of the Yunnan Institute of Tropical Crops (http://www.yitc.com.cn, Dr. Jin Liu, [email protected]), voucher numbers YITC-2020-FZ-E-115 and D2020-FZ-E-115, respectively.
The total DNA was used to produce paired-end (PE) Illumina sequencing libraries with 350 bp average insert size, and sequenced on the Illumina HiSeq 2500 platform (Illumina, San Diego, CA). The generated 7.5 Gb raw data were filtered and assembled using the SPAdes-3.5.0 (http://soap.genomics.org.cn/soapdenovo.html), following the sequence overlap and PE relationships. Sanger sequencing was applied to verify the four boundaries of the IR region and the chloroplast genome was annotated using CpGAVAS2 (Shi et al. Citation2019) and GeSeq (Tillich et al. Citation2017). The complete, annotated chloroplast genome was submitted to GenBank (http://www.ncbi.nlm.nih.gov/), accession number MW528030.
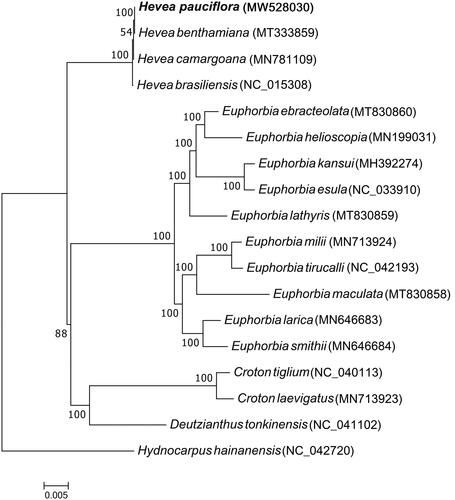
Next, the H. pauciflora chloroplast genome was phylogenetically analyzed using the maximum-likelihood method and 17 () other Euphorbiaceae species to understand the phylogenetic relationship between the 18 chloroplast genomes. The 17 Euphorbiaceae species included four genera: Euphorbia (10 species), Hevea (four species), Croton (two species), and Deutzianthus (one species). Hydnocarpus hainanensis, a tree species belonging to the Achariaceae family (order Malpighiales, as with the other 17 species), was the outgroup. Multiple sequence alignment was performed using MAFFT (Katoh and Standley Citation2013), whereas RAxML8.2.4 was employed to conduct phylogenetic analysis (Stamatakis Citation2014). Node support was estimated from the results of 1000 bootstrap replicates.
Figure 2. Maximum-likelihood tree based on complete chloroplast genome sequences of H. pauciflora and other Euphorbiaceae species. Hydnocarpus hainanensis, a tree species belonging to the Achariaceae family, was the outgroup.
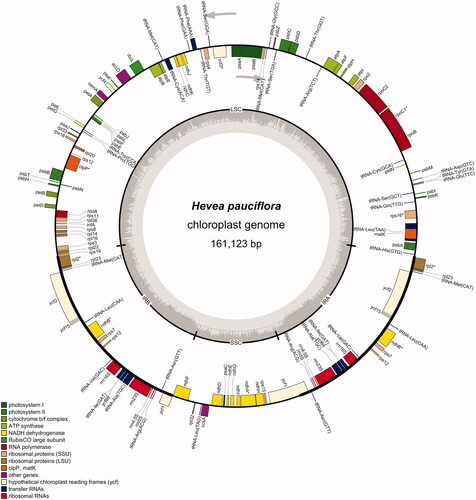
The complete chloroplast genome of H. pauciflora is 161,123 bp long, with a canonical quadripartite structure (). The structure contains a large single-copy (LSC) region of 89,109 bp (33.18% GC content), a small single-copy (SSC) region of 18,376 bp (29.42% GC content), and two inverted repeat regions (IRa and IRb) of 26,819 bp (42.20% GC content). Furthermore, the annotated H. pauciflora chloroplast genome contains 134 genes, including 86 protein-coding genes, 36 tRNA genes, eight rRNA genes, and four pseudogenes. The whole chloroplast genome contains 50,494 A bases (31.96%), 52,023 T bases (32.29%), 28,915 G bases (17.95%), and 28,691 C bases (17.81%). Moreover, the 134 genes contain four gene categories: self-replication, photosynthesis, unknown function, and other genes.
Phylogenetic analysis showed that H. pauciflora, H. brasiliensis, H. camargoana, and H. benthamiana are closely clustered in one clade (), consistent with traditional taxonomy. This H. pauciflora chloroplast genome sequence provides useful data for further studies of the Hevea genus and understanding the phylogenetic relationships of Euphorbiaceae species.
Ethics statement
The collection of specimens conformed to the requirement of international ethics, which did not cause damage to the local environment. The process and purpose of this experimental research were in line with the rules and regulations of our institute. There are no ethical issues or other conflicts of interest in this study.
Author contributions
Hua-Sun Huang and Jin Liu conceived the study, wrote and revised the manuscript. Yan-Shi Hu collected and managed the experimental material plants. Jin Liu and Yan-Shi Hu performed the data analyses and drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. MW528030. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA763296, SRR15911750, and SAMN21437637, respectively.
Additional information
Funding
References
- Baldwin JJT. 1947. Hevea: a first interpretation. A cytogenetic survey of a controversial genus, with a discussion of its implications to taxonomy and to rubber production. J Hered. 38(2):54–64.
- Clément-Demange A, Priyadarshan PM, Thuy Hoa TT, Venkatachalam P. 2007. Hevea rubber breeding and genetics. In: Janick J, editor. Plant breeding reviews. Vol. 29. Hoboken: John Wiley & Sons, Inc.; p. 177–283.
- Goncalves P, Cardoso M, Ortolani AA. 1990. Origin, variability and domestication of Hevea. Pesq Agropec Bras. 25:135–156.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lau NS, Makita Y, Kawashima M, Taylor TD, Kondo S, Othman AS, Shu-Chien AC, Matsui M. 2016. The rubber tree genome shows expansion of gene family associated with rubber biosynthesis. Sci Rep. 6:28594.
- Liu J, Shi C, Shi CC, Li W, Zhang QJ, Zhang Y, Li K, Lu HF, Shi C, Zhu CS, et al. 2020. The chromosome-based rubber tree genome provides new insights into spurge genome evolution and rubber biosynthesis. Mol Plant. 13(2):336–350.
- Majumder SK. 1964. Chromosome studies of some species of Hevea. J Rubb Res Inst Malaysia. 18:269–273.
- Niu YF, Hu YSH, Zheng C, Liu ZY, Liu J. 2020. The complete chloroplast genome of Hevea camargoana. Mitochondrial DNA Part B. 5(1):607–608.
- Niu YF, Hu YSH, Liu ZY, Zheng C, Liu J. 2020. Complete chloroplast genome of Hevea benthamiana, a SALB-resistant relative wild species of rubber tree. Mitochondrial DNA Part B. 5(3):2062–2064.
- Pootakham W, Sonthirod C, Naktang C, Ruang-Areerate P, Yoocha T, Sangsrakru D, Theerawattanasuk K, Rattanawong R, Lekawipat N, Tangphatsornruang S. 2017. de novo hybrid assembly of the rubber tree genome reveals evidence of paleotetraploidy in Hevea species. Sci Rep. 7:41457.
- Priyadarshan PM, Goncalves P. 2002. Use of Hevea gene pool in rubber tree (Hevea brasiliensis Muell Arg.) breeding. Planter. 78:123–138.
- Rahman AYA, Usharraj AO, Misra BB, Thottathil GP, Jayasekaran K, Feng Y, Hou SB, Ong SY, Ng FL, Lee LS, et al. 2013. Draft genome sequence of the rubber tree Hevea brasiliensis. BMC Genomics. 14(1):75.
- Shi LC, Chen HM, Jiang M, Wang LQ, Wu X, Huang LF, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tang CR, Yang M, Fang YJ, Luo YF, Gao SH, Xiao XH, An ZW, Zhou BH, Zhang B, Tan XY, et al. 2016. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat Plants. 2(6):16073.
- Tangphatsornruang S, Uthaipaisanwong P, Sangsrakru D, Chanprasert J, Yoocha T, Jomchai N, Tragoonrung S. 2011. Characterization of the complete chloroplast genome of Hevea brasiliensis reveals genome rearrangement, RNA editing sites and phylogenetic relationships. Gene. 475(2):104–112.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.