Abstract
Suaeda physophora Pall. (Chenopodiaceae) is a leaf succulent shrub species with potential usefulness as fodder for the desert animal. However, the phylogeny of S. physophora is lacking. Here, we sequenced and assembled a complete chloroplast genome of S. physophora and further reconstructed the phylogeny of Chenopodiaceae. The chloroplast genome of S. physophora is 151,104 bp in length, consisting of an 18,597 bp small single-copy (SSC), an 82,845 bp large single-copy (LSC), and a pair of 24,831 bp inverted repeat (IR) regions. The genome encodes 131 genes, including 87 protein-coding genes, 36 tRNA genes, and eight rRNA genes. Phylogenetic analysis revealed that the genus Suaeda forms a monophyletic taxon, and S. physophora is closely related to S. eltonica. Chloroplast genome and phylogenetic studies provided an essential foundation for the conservation of S. physophora.
Keywords:
Suaeda physophora Pall. (1803) is a salt-resistant halophyte distributed in northwestern China, Eastern Europe, Central Asia, and Western Siberia (Song et al. Citation2005). Owing to its importance as fodder in areas with saline soil, the salt-tolerance mechanisms of S. physophora have been extensively studied (Song et al. Citation2006; Li and Zhang Citation2007; Yuan et al. Citation2010; Yang et al. Citation2016). However, the phylogeny of this species is not comprehensively resolved. A previous study used one nuclear locus, two plastid loci, and morphological characteristics to infer the phylogeny and taxonomy of Chenopodiaceae (Schütze et al. Citation2003); as only atpB-rbcL reference sequences were available, S. physophora was grouped together with S. palaestina and S. ifniensis; however, their phylogenetic positions were not yet resolved. Thus, more comprehensive and stronger molecular evidence is still needed, especially at the plastomic level. To address this, we assembled and decoded the chloroplast genome of S. physophora, representing the first complete chloroplast genome of this species.
We collected samples of S. physophora growing wild in Xinjiang, China (42.8488° N, 89.1975° E), in accordance with local laws and exerting no environmental damage. Species identification was based on morphology and subsequent chloroplast genome sequencing. The S. physophora specimen was deposited at the Molecular Evolution and Ecology Laboratory, Fudan University (contact: Jiayin Zhang, email: [email protected]), voucher Sa004-HP002-Sphy. Total genomic DNA was extracted using Plant DNAzol Reagent (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer`s instructions and was sequenced on an Illumina Hiseq 2500 platform (Illumina, San Diego, CA) by a commercial service provider (BerryGenomics, Beijing, China) in 150-bp paired-end sequencing mode. The S. physophora chloroplast genome was assembled using GetOrganelle v1.7.5 (Jin et al. Citation2020), followed by manual correction using Bandage v0.8.1 (Wick et al. Citation2015). The final assembly was curated using Pilon v1.22 (Walker et al. Citation2014), and annotation was performed on GeSeq (Tillich et al. Citation2017) with all available chloroplast genomes of Suaeda (Chenopodiaceae) as references. The assembled genome sequence and annotations were submitted to GenBank (accession no. ON571659).
The assembled S. physophora chloroplast genome was 151,104 bp long, including an 18,597-bp small single-copy (SSC), an 82,845-bp large single-copy (LSC), and a pair of 24,831-bp inverted repeat (IR) regions. We annotated 131 genes, including 87 protein-coding genes, eight rRNA genes, and 36 tRNA genes, among which 10 tRNA genes, four rRNA genes, and eight protein-coding genes were duplicate in IR regions. The average GC content of the LSC, SSC, and IR regions is 34.3%, 29.0%, and 42.8%, respectively.
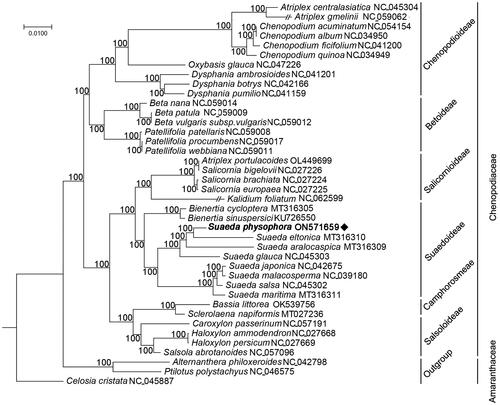
To resolve the taxonomic status at plastomic level, the complete chloroplast genomes of S. physophora and 38 other species belonging to the Chenopodiaceae and Amaranthaceae were used to construct a phylogenetic tree. MAFFT v7.487 (Rozewicki et al. Citation2019) and TrimAl v1.4.rev22 (Capella-Gutiérrez et al. Citation2009) were used to align and trim the genome sequences. We used IQ-TREE v2.1.2 (Minh et al. Citation2020) to construct a maximum-likelihood tree with a TVM + F+R4 model. Phylogenetic analysis results () strongly suggested that the genus Suaeda is monophyletic, and S. physophora is a sister species to S. eltonica.
Author contributions
Y.H. and J.Z. designed the study. Y.H. and X.R. collected plant samples, performed bioinformatics analysis, and wrote the manuscript. J.Z. revised the manuscript. All the authors approved the final manuscript text.
Disclosure statement
In accordance with Taylor & Francis policy and our ethical obligation as a researcher, the authors are reporting that Y.H. and X.R. have a business interest in a company that may be affected by the research reported in the enclosed paper. The authors have disclosed those interests fully to Taylor & Francis, and they have in place an approved plan for managing any potential conflicts arising from that involvement. No other potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. ON571659. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA844225, SRR19501374, and SAMN28798005, respectively.
References
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Li L, Zhang X. 2007. Germination strategies of two halophytes in Salt Desert of northwestern China. Sci China Ser D. 50(S1):115–121.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Rozewicki J, Li S, Amada KM, Standley DM, Katoh K. 2019. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 47(W1):W5–W10.
- Schütze P, Freitag H, Weising K. 2003. An integrated molecular and morphological study of the subfamily Suaedoideae Ulbr. (Chenopodiaceae). Plant Syst Evol. 239(3–4):257–286.
- Song J, Ding X, Feng G, Zhang F. 2006. Nutritional and osmotic roles of nitrate in a euhalophyte and a xerophyte in saline conditions. New Phytol. 171(2):357–365.
- Song J, Feng G, Tian CY, Zhang FS. 2005. Strategies for adaptation of Suaeda physophora, Haloxylon ammodendron and Haloxylon persicum to a saline environment during seed-germination stage. Ann Bot. 96(3):399–405.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9(11):e112963.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Yang X, Yu H, Zhang T, Guo J, Zhang X. 2016. Arbuscular mycorrhizal fungi improve the antioxidative response and the seed production of Suaedoideae species Suaeda physophora Pall under salt stress. Notul Bot Horti Agrobot. 44(2):533–540.
- Yuan J-F, Feng G, Ma H-Y, Tian C-Y. 2010. Effect of nitrate on root development and nitrogen uptake of Suaeda physophora under NaCl salinity. Pedosphere. 20(4):536–544.