Abstract
Cnaphalocrocis patnalis (Bradley 1981) is a major pest that threatens the safety of rice production in the world. In the present study, we determined the complete mitogenome of C. patnalis. This mitogenome was 15,305 bp in length (GenBank accession No. OL449028), which contained two ribosomal RNA genes, 22 transfer RNAs, 13 protein-coding genes (PCGs) and one non-coding AT-rich region with a length of 344 bp. All the 22 tRNA genes displayed a typical clover-leaf structure, except for trnS1. Twelve PCGs were initiated by ATN codons, and COX1 started with TTG. All the PCGs used the typical stop codon ‘TAA’ and ‘TAG.’ Phylogenetic tree demonstrated that C. patnalis belongs to the family Crambidae.
The rice leaf folder, Cnaphalocrocis patnalis Bradley (Lepidoptera: Crambidae) is a worldwide pest that damages rice through rolling and feeding with the rice leaves (Barrion et al. Citation1991). The leaf folder has caused up to 57% of the expected crop yield loss (Karim and Dean Citation2000). To date, research on this pest has focused on biological characteristics and patterns of occurrence, while studies on its evolutionary history have not been reported. In this study, we sequenced and annotated the complete mitogenome of C. patnalis.
Here, we sequenced and annotated the complete mitogenomes of C. patnalis, and created the phylogenetic tree for taxonomy classification. Individuals of C. patnalis were collected from Yazhou District in Sanya (18°19′22″N, 109°10′52″E), Hainan, China. All insect handling and experimental procedures were approved by the Ethics Committee of Chinese Academy of Tropical Agricultural Sciences (Hainan, China). The specimens were stored at −80 °C in the Environment and Plant Protection Institute, Chinese Academy of Tropical Agricultural Sciences, Haikou, China (Jihong Tang, [email protected]), under the accession number IN07040201-0001-00025. Total genomic DNA was extracted from single sample by the CTAB method (Reineke et al. Citation1998). The mitochondrial genome sequence was generated using Illumina HiSeq X TEN Sequencing System with 150 bp paired-end reads. The sequence was assembled by the MITObim software (Hahn et al. Citation2013) and checked by Geneious Primer (http://www. geneious.com/). The annotation of the mitochondrial genome sequence was mainly compared with the existing mitochondrial genomes of related species, and the annotation results were confirmed by the MITOS web server (Bernt et al. Citation2013). The sequence was submitted to GenBank under the accession number OL449028.
The C. patnalis mitogenome was 15,305 bp in length consisting of 13 PCGs, 22 tRNAs, two rRNAs, and a non-coding AT-rich region with a length of 344 bp. The nucleotide composition of C. patnalis mitogenome was biased toward AT at 96.2%, and the total base composition was 41.3% A, 54.9% T, 0.6% G, and 3.2% C. Twenty-two genes were encoded on the majority (J) strand, while 15 genes were encoded on minority (N) strand. Twelve PCGs started with ATN, except that COX1 was initiated with TTG. Eleven PCGs used the typical stop codon ‘TAA,’ NAD2 and NAD5 used ‘TAG.’ The length of tRNA genes ranges from 48 to 72 bp and all of them can be folded into the typical clover-leaf secondary structure, except for trnS1, which lack dihydrouridine (DHU) arm. The 16S rRNA was 1350 bp long with an AT content of 85.2%, while the 12S rRNA was 779 bp long with an AT content of 86.5%.
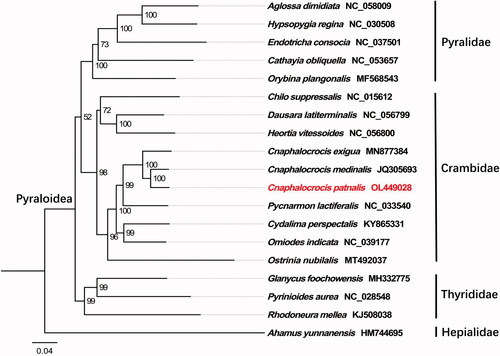
Phylogenetic relationship of C. patnalis based on the concatenated datasets of the 13 PCGs from 17 Pyraloidea species was carried out, using Ahamus yunnanensis (Hepialoidea: Hepialidae) as an outgroup. The analyses were performed by the maximum likelihood using PhyloSuite v1.2.2 with 5000 bootstrap replicates (Zhang et al. Citation2020). FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/) to visualize the phylogenetic tree. The phylogenetic tree () demonstrated C. patnalis belongs to the family Crambidae, which accord with the conventional classification. In conclusion, the results of this study provide an essential DNA molecular database for further evolutionary and molecular research on Crambidae.
Author contributions
Design of the work: Baoqian Lyu, Jinhua Li and Xue Tang. Performed the experiments: Nanfang Lin, Chunxi Cheng and Jinhua Li. Interpretation of data for the work: Hui Lu, Jihong Tang . Wrote and revised of the paper: Jihong Tang, Baoqian Lyu, Xue Tang and Hui Lu. All authors agree to be accountable for all aspects of the work.
Acknowledgments
We thank Baoqian Lyu for his help collecting the described samples of the insects. We gratefully acknowledge the financial contribution to this project by the National Key Research and Development Project (2019YFD03001 and 2019YFD1002100).
Disclosure statement
We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession No. OL449028. The associated BioProject, Bio-Sample and SRA numbers are PRJNA780076, SAMN23101374 and SRR16938303, respectively.
Additional information
Funding
References
- Barrion AT, Litsinger JA, Medina EB, Aguda RM, Bandong JP, Pantua PC, Viajante VD, Jr, Dela Cruz CG, Vega CR, Soriano JS, et al. 1991. The rice Cnaphalocrocis and Marasmia (Lepidoptera: Pyralidae) leaffolder complex in the Philippines: taxonomy, bionomics, and control. Philipp Ent. 8(4):987–1074.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Karim S, Dean DH. 2000. Toxicity and receptor binding properties of Bacillus thuringiensis delta-endotoxins to the midgut brush border membrane vesicles of the rice leaf folders, Cnaphalocrocis medinalis and Marasmia patnalis. Curr Microbiol. 41(4):276–283.
- Reineke A, Karlovsky P, Zebitz C. 1998. Preparation and purification of DNA from insects for AFLP analysis. Insect Mol Biol. 7(1):95–99.
- Zhang D, Gao FL, Li WX, Jakovlić I, Zou H, Zhang J, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.