Abstract
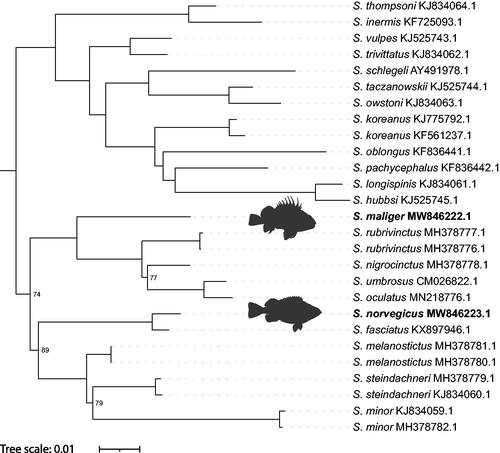
We report the complete mitochondrial genomes of two rockfish: Sebastes maliger and Sebastes norvegicus. The mitogenomes consist of 13 protein-coding regions, 22 tRNAs, two rRNAs, and one control region. Sebastes mitogenome control regions are highly variable due to the presence of repeat sequences. The mitogenomes for S. maliger and S. norvegicus are 16,403 and 16,401 bp, respectively. Using these two mitogenomes and 25 additional Sebastes mitogenomes from GenBank, we examine the phylogenetic relationships in Sebastes. Sebastes maliger is sister to a clade including S. rubrivinctus, S. nigrocinctus, S. umbrosus, and S. oculatus, while S. norvegicus is sister to S. fasciatus.
Sebastes (Cuvier, 1829) is a diverse genus of marine fish, comprising at least 110 species found mostly in the North Pacific, with a small number of species in the Atlantic, Arctic, and Indian Oceans (Kim and Lee Citation2004; Hyde and Vetter Citation2007). The Quillback rockfish, Sebastes maliger (Jordan and Gilbert, 1880), is found in the North-East Pacific Ocean (Munk Citation2001; Love et al. Citation2002; West et al. Citation2014); whereas the Golden redfish, Sebastes norvegicus (Ascanius, 1772), is found in the North Atlantic Ocean (Rolskii et al. Citation2020). Although previous studies have focused on the mitogenomes of Western Pacific rockfish, few studies have evaluated the mitogenomes of East Pacific and Atlantic species. We provide further resolution of the phylogenetic relationships in Sebastes by assembling the complete mitogenomes of S. maliger and S. norvegicus.
We collected S. maliger near Excursion Inlet, Alaska in the Gulf of Alaska (58.4854, −135.4521) using hook-and-line sampling (IACUC-approved protocol #15-0602). A liver sample was placed in RNAlater (MilliporeSigma, St. Louis, MO, USA), frozen at −20 °C, transported to Brigham Young University, and stored at −80 °C until processed. A morphological voucher was not retained. We extracted DNA and sequenced it using an Illumina HiSeq 2500 system (Illumina, San Diego California, USA) at Brigham Young University’s DNA Sequencing Center (Provo, Utah, USA). The tissue sample (267106) was deposited at the Monte L. Bean Life Science Museum, Brigham Young University (Jerald B. Johnson, [email protected]) and raw sequences were deposited in the SRA database (SRR15573079). We retrieved DNA sequences of S. norvegicus (ERR1473911) from the SRA database on NCBI (Malmstrøm et al. Citation2017). This specimen was collected near Ballstad in Lofoten, Norway (68.0713, 13.5361) by commercial fishermen (M. Malmstrøm, personal communication, November 29, 2021).
We checked sequence quality using FastQC (Andrews Citation2010). Mitogenomes were assembled with Geneious v. 11.0.9 (Biomatters Ltd., Auckland, New Zealand) by mapping short reads against three reference mitogenomes, S. fasciatus, S. oculatus, and S. koreanus, (KX897946, MN218776, KJ775792). We then extracted a single consensus contig for each assembly (>100x coverage for both mitogenomes for all assemblies). The mitogenomes were annotated using MitoAnnotator (Iwasaki et al. Citation2013). For our phylogenetic analysis, we acquired a total of 25 rockfish mitogenomes from GenBank. We generated multiple sequence alignments separately for each of the 13 protein-coding genes using MAFFT v. 7.475 (Katoh and Standley Citation2013) before concatenating them into a final file that was 11,412 bp in length after all positions with missing data were removed.
The complete mitogenomes of S. maliger and S. norvegicus, assembled against S. fasciatus (KX897946), were 16,403 and 16,401 bp in length, respectively (MW846222, MW846223). Depending on which reference mitogenome was used during assembly (KX897946, MN218776, KJ775792), the complete mitogenomes of S. maliger and S. norvegicus were between 16,401 and 17,609 bp, consistent with studies showing that the mtDNA control region of Sebastes is highly variable due to repetitive DNA sequences (Zhang et al. Citation2013; Sandel et al. Citation2018). The mitogenomes consist of 13 protein-coding genes, 22 tRNAs, two rRNAs, and one control region, and the order is identical to published Sebastes mitogenomes (Kim and Lee Citation2004; Zhang et al. Citation2013; Jang et al. Citation2016; Oh et al. Citation2016; Sandel et al. Citation2018; Kim et al. Citation2019). Twelve of the 13 protein coding genes had an ATG start codon; COX1 had a GTG start codon. Six genes (ND1, COX1, ATP8, ND4L, ND5, and ND6) had a TAA stop codon, three genes (ND2, ATP6, and COX3) had an incomplete TA stop codon, and four genes (COX2, ND3, ND4, and CytB) had an incomplete T stop codon.
Two phylogenetic trees were generated using W-IQ-Tree (Trifinopoulos et al. Citation2016) and BEAST 2.5 (Bouckaert et al Citation2019). We used Sebasticus tertius (MT117231) as an outgroup. Both phylogenetic analyses indicated that S. maliger is sister to a clade including S. rubrivinctus, S. nigrocinctus, S. umbrosus, and S. oculatus, and that S. norvegicus is sister to S. fasciatus (). For the maximum likelihood phylogeny, all relationships are consistent with those previously published, except for S. steindachneri, which is sister to S. minor instead of S. melanostictus (Sandel et al. Citation2018; Kim et al. Citation2019), and S. owstoni, which is sister to S. taczanowskii instead of S. minor (Hyde and Vetter Citation2007). The Bayesian phylogeny (not shown) is identical to the maximum likelihood phylogeny except that S. steindachneri is sister to S. melanostictus instead of S. minor, and S. nigrocinctus is sister to S. rubrivinctus instead of S. umbrosus and S. oculatus. Several nodes in our maximum likelihood phylogeny had low bootstrap support (≤95) while all nodes in the Bayesian phylogeny had posterior probabilities >.99. Although additional species and analyses are needed to explore the differences seen in our phylogenies, the mitogenomes of S. maliger and S. norvegicus provide new insight into the phylogenetic relationships in Sebastes.
Figure 1. Phylogenetic tree inferred by maximum likelihood using W-IQ-Tree from 27 Sebastes mitogenomes. Sebastiscus tertius (MT117231) was used as an outgroup but is not displayed. Ultrafast bootstrap values >95 are not shown. “Sebastes norvegicus” by Jan Fekian is licensed under CC BY-SA 4.0/Silhouette of original.
Ethical approval
Sebastes maliger was collected under Brigham Young Universities’ IACUC-approved protocol #15-0602 for DKS. Sebastes norvegicus was caught by commercial fisherman (Malmstrøm et al. Citation2017).
Author contributions
The author contributions were as follows: Jillian R. Campbell: Analysis and Interpretation, Writing – Original Draft, Writing – Review and Editing., Peter C. Searle: Conception and Design, Analysis and Interpretation, Writing – Review and Editing. Andrea L. Kokkonen: Conception and Design, Analysis and Interpretation, Writing – Review and Editing. Dennis K. Shiozawa: Conception and Design, Analysis and Interpretation, Writing – Review and Editing. Mark C. Belk: Conception and Design, Analysis and Interpretation, Writing – Review and Editing. R. Paul Evans: Conception and Design, Analysis and Interpretation, Writing – Review and Editing. All authors approve the publication of this manuscript and agree to be accountable for all aspects of the work.
Acknowledgements
We thank Scott and Jody Jorgenson at Pybus Point Lodge, the Roger and Victoria Sant Foundation, the College of Life Sciences at Brigham Young University, the Department of Biology at Brigham Young University, and the Monte L. Bean Life Science Museum for supporting this study.
Disclosure statement
The authors declare no potential conflict of interest, and the authors alone are accountable for the content and composition of the paper.
Data availability statement
The genome sequence data that support the findings of this study are publically available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under accession numbers MW846222 and MW846223. The associated BioProject number for S. maliger and S. norvegicus is PRJNA741690, which holds a growing dataset of Sebastes mitogenomes. The associated BioSample numbers for S. maliger and S. norvegicus are SAMN20892470 and SAMEA4028819, respectively. The associated SRA numbers for S. maliger and S. norvegicus are SRR15573079 and ERR1473911, respectively.
Additional information
Funding
References
- Andrews S. 2010. FastQC: A quality control tool for high throughput sequence data [Online]. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N, et al. 2019. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 15(4):e1006650.
- Hyde JR, Vetter RD. 2007. The origin, evolution, and diversification of rockfishes of the genus Sebastes (Cuvier). Mol Phylogenet Evol. 44(2):790–811.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540.
- Jang YS, Park KJ, Kim KY, Kim S. 2016. The complete mitochondrial genome of Sebastes pachycephalus (Scorpaenidae, Scorpaeniformes) from the East Sea, Korea. Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):69–70.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim IC, Lee JS. 2004. The complete mitochondrial genome of the rockfish Sebastes schlegeli (Scorpaeniformes, Scorpaenidae). Mol Cells. 17(2):322–328.
- Kim H, Yoon M, Kim HJ. 2019. The complete mitochondrial genome of rockfish Sebastes oculatus Valenciennes, 1833 from southwest Atlantic ocean. Mitochondrial DNA B Resour. 4(2):3407–3408.
- Love MS, Yoklavich M, Thorsteinson L. 2002. The rockfishes of the Northeast Pacific. Berkeley: University of California Press.
- Malmstrøm M, Matschiner M, Tørresen OK, Jakobsen KS, Jentoft S. 2017. Whole genome sequencing data and de novo draft assemblies for 66 teleost species. Sci Data. 4:160132.
- Munk KM. 2001. Maximum ages of groundfishes in waters off Alaska and British Columbia and considerations of age determination. Alaska Fish Res Bull. 8:12–21.
- Oh J, Kim S, Lee EK, Park JH, Kim KY, Jang YS. 2016. Complete mitochondrial genome of Sebastes owstoni (Scorpaenidae, Scorpaeniformes) from the East Sea, Korea. Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):3952–3954.
- Rolskii AY, Artamonova VS, Makhrov AA. 2020. Molecular identification of golden redfish (Sebastes norvegicus) in the White Sea. Polar Biol. 43(4):385–389.
- Sandel MW, Aguilar A, Buonaccorsi VP, Herstein J, Evgrafov O. 2018. Complete mitochondrial genome sequences of five rockfishes (Perciformes: Sebastes). Mitochondrial DNA B Resour. 3(2):825–826.
- Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.
- West JE, Helser TE, O'Neill SM. 2014. Variation in quillback rockfish (Sebastes maliger) growth patterns from oceanic to inland waters of the Salish Sea. BMS. 90(3):747–761.
- Zhang H, Zhang Y, Zhang X, Song N, Gao T. 2013. Special structure of mitochondrial DNA control region and phylogenetic relationship among individuals of the black rockfish, Sebastes schlegelii. Mitochondrial DNA. 24(2):151–157.
