Abstract
Monoon laui (Merr.) B. Xue and R.M.K. Saunders 2012 is produced in Hainan province. The trunk is straight, the wood texture is straight, and the material is slightly soft, which is suitable for furniture and building materials. In our study, we report and characterize the complete plastome of M. laui The complete length of the plastome of M. laui possesses 161,181 bp, including a large single-copy (LSC) of 89,556 bp, small single-copy (SSC) of 18,977 bp, and two inverted repeats (IRs) of 26,313 bp. The overall G/C content in the plastome of M. laui is 39.13%. The plastome contains 257 genes, consisting of 130 protein-coding genes (16 of which are duplicated in the IR), 37 tRNA genes (seven of which are duplicated in the IR), and eight rRNA genes (5S rRNA, 4.5S rRNA, 16S rRNA, and 23S rRNA). Here, we explore the phylogenetic relationships and make contributions to the conservation genetics of the specie of M. laui using the complete plastome sequence.
Introduction
Monoon laui (Merr.) B. Xue and R.M.K. Saunders 2012. belong to the family Annonaceae Juss. M. laui is a tree. The leaves are nearly leathery to leathery, oblong or oblong-elliptic, 8–20 cm long, 3.5–8 cm wide, acuminate at the top, broad and acute or rounded at the base, glabrous and shiny on both sides. It is produced in the low to mid-altitude forests of Hainan province. The trunk is straight, the bark is yellowish brown, and the flowers are pale yellow, which can be used as a greening and beautifying tree species for gardens and sidewalks. The wood is straight and slightly soft, suitable for furniture and building materials. At present, the systematic position of M. laui have not been reported. Hence, we used the complete plastid of M. laui (GenBank accession number: OL979152, this study) to explore the phylogenetic relationships, in order to facilitate the collection, preservation and systematic research of M. laui germplasm resources.
In this study, M. laui was sampled from Ledong county in Hainan province of China (108.79° E, 18.69° N). The leaves were deposited in silica gel. DNA was extracted from dried leaf tissue using a modified cetyltrimethylammonium bromide (CTAB) method, with chloroform: isoamyl alcohol separation and isopropanol precipitation at −20 °C. Its DNA was deposited in the Herbarium of the Institute of Tropical Agriculture and Forestry (code of herbarium: HUTB), Hainan University, Haikou, China.
The experiment procedure is as reported in Wang et al. (Citation2021). Each individual is based on 6 Gb clean data, adapter sequences and low-quality reads with Q-value ≤20 were removed. Clean reads were assembled against the plastome of Polyalthiopsis verrucipes (MW018366.1) using MITObim v.1.8 (Hahn et al. Citation2013). The plastome was annotated using Geneious R8.0.2 (Biomatters Ltd., Auckland, New Zealand).
The result of our study indicates that the complete length of the plastome of M. laui possesses 161,181 bp with the typical quadripartite structure of angiosperms. This plastid contains an LSC region of 89,556 bp and an SSC region of 18,977 bp, two IRs of 26,313 bp. The plastome contains 257 genes, consisting of 130 protein-coding genes (sixteen of which are duplicated in the IR), 37 tRNA genes (seven of which are duplicated in the IR), and eight rRNA genes (5S rRNA, 4.5S rRNA, 16S rRNA, and 23S rRNA). The overall G/C content in the plastome of M. laui is 39.13%, while the corresponding value of the LSC, SSC, and IR regions were 37.7%, 34.3%, and 43.2%, respectively.
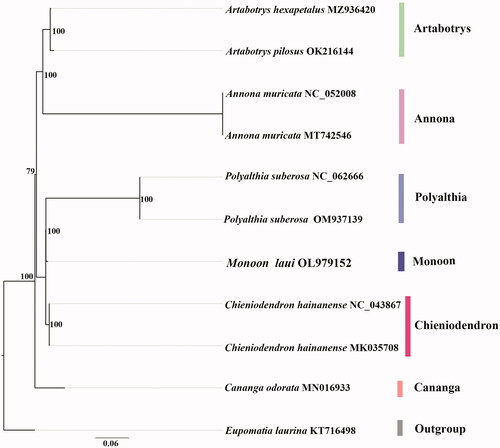
We used IQ-TREE v.1.6.1 (Nguyen et al. Citation2015) to establish evolutionary relationships (Kalyaanamoorthy et al. Citation2017). We inferred the phylogeny of M. laui based on plastid alignments (). The majority of nodes in the plastome ML trees were highly supported. The phylogenetic analysis indicates that M. laui is sister to the clade of Polyalthia suberosa (Roxb.) Thwaites having strong support ().
Figure 1. The best ML phylogeny recovered from 11 complete plastome sequences by RAxML. Accession numbers: Monoon laui (GenBank accession number, OL979152, this study), Artabotrys hexapetalus, MZ936420, Artabotrys pilosus, OK216144, Annona muricata, MT742546, Annona muricata, NC_052008, Polyalthia suberosa, OM937139, Polyalthia suberosa, NC_062666, Chieniodendron hainanense, NC_043867, Chieniodendron hainanense, MK035708, Cananga odorata, MN016933, Eupomatia laurina, KT716498.
Nowadays, the plastid sequence of M. laui has been gradually developed and tended to be perfect and phylogenetic studies of M. laui can be explored more sufficiently.
Author contributions
Qinghui Sun involved in the conception and design. Hao Xiu, Yunfan Quan, Tingting Fu analyzed and interpreted the data; Hao Xiu, Yunfan Quan, Tingting Fudrafted the paper. Qinghui Sun revised it critically for intellectual content. All authors final approved the version to be published. All authors agreed to be accountable for all aspects of the work.
Ethics statement
The study was approved by the institutional review board of Hainan Medical University, Haikou, China. The collection of plant materials is carried out in accordance with guidelines provided by the Hainan Medical University and Hainan province regulations. Field studies comply with Hainan province field work policy drafted by Hainan Medical University (in Chinese), and the manuscript include a statement of appropriate permissions granted and licenses from related agencies of Hainan province. Voucher specimens are deposited in a public herbarium (HUTB) providing access to deposited material. Information on the voucher specimen and who identified it is included in the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data supporting the results of this study are publicly trustworthy in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/)with registration number OL979152. Related BioProject, SRA, Biosample numbers are PRJNA748537, SRS12065959, and SAMN20703218, respectively. A specimen was deposited at Hainan University (https://ha.hainanu.edu.cn/home2020/,Qinghui Sun and [email protected]) under the voucher number A209.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads: a baiting and iterative mapping approach. Nucleic Acids Research. 41(13):e129.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Wang HX, Morales-Briones DF, Moore MJ, Wen J, Wang HF. 2021. A phylogenomic perspective on gene tree conflict and character evolution in Caprifoliaceae using target enrichment data, with Zabelioideae recognized as a new subfamily. J Syst Evol. 59(5):897–914.
