Abstract
Recently, the chloroplast genome of Viola verecunda from a sample collected in Japan has been published. Although the name is often recognized as a taxonomic synonym of Viola arcuata, the genetic identity of the two species has never been compared intensively. We report the complete chloroplast genome sequence of V. arcuata, of which sample was collected from Seoul, Korea. The cp genome of V. arcuata (OM301625) has 157,870 bp in length and is composed of four regions: 86,366 bp of a large single-copy (LSC) region, 17,298 bp of a small single-copy (SSC) region, and 27,103 bp of a pair of inverted repeats (IRs). The complete genome contains 130 genes, including 84 protein-coding genes, eight rRNA genes, and 37 tRNA genes. When comparing chloroplast genomes between V. verecunda, and V. arcuata, 34 different loci were recognized: 12 SNPs and 22 indels. In the coding regions, there were two amino acid insertions (ndhI) caused by one base deletion, three synonymous substitutions (ndhF, ccsA, and ndhI), and six nonsynonymous substitutions (matK, rpoC2, ndhF, ycf1, and two rpl2s on each IR region). In non-coding regions, variants of 19 polyN sites, one microsatellite, two insertions, and two SNPs were recognized. Phylogenetic analysis confirms a sister or nearly identical relationship between two genomes. This study will provide the genetic basis for solving a taxonomic problem between V. arcuata and V. verecunda.
Viola L., the largest genus in Violaceae Batsch, contains 583–620 species distributed worldwide (Wahlert et al. Citation2014, Cheon et al. Citation2019). Viola arcuata Blume Citation1825 included in the subsection Bilobatae of section Plagiostigma (Marcussen et al. Citation2022) in Viola is originally described with a collection from Java (Blume Citation1825) and is widely distributed in Asia and Russia (Chen et al. Citation2007). Viola verecunda A. Gray Citation1857, of which type specimen is collected in Japan (Gray Citation1857), is often recognized as a taxonomic synonym of V. arcuata (Chang et al. Citation2014) or Viola hamiltoniana D. Don et al. Citation1825, which is described with a specimen collected in Nepal (Don et al. Citation1825). Although these species have some shared morphology in type specimens (e.g. distinct stems, ovate-cordite leaves, and white flowers with small spurs), extensive variations in leaf shape and size have been observed in the field. All of these names are still widely used in recent floral works and phylogenetic studies (Juzepchuk and Klokov Citation1949; Akiyama et al. Citation2002; Chen et al. Citation2007; Yoo and Jang Citation2010; Chayamarit and Balslev Citation2018; Lee and Yoo Citation2020) without intensive morphological and genetic investigations.
Recently, the importance of the taxonomic entity of V. verecunda has been emphasized because of the usage of this species. The triterpenoids of this plant exhibit high antiplasmodial activity against malaria (Moon et al. Citation2007). Furthermore, plant extracts affect hair loss prevention in humans (Kang et al. Citation2021). Therefore, it is necessary to clarify the taxonomic identity of this species through morphological and genetic studies. In this study, we report the complete chloroplast (cp) genome of V. arcuata using a sample collected in the Korean peninsula and compare it with previously reported that of V. verecunda, which the sample collected in Japan (Kwak Citation2021).
The sample was collected at Mia-dong, Gangbuk-gu, Seoul, the Republic of Korea (N 37°37′54.84″, E 127°01′36.07″). The voucher specimen (H. Moon 2021-46) was deposited in the herbarium of Sungshin Women’s University (SWU; Sangtae Kim, [email protected]). Total genomic DNA was extracted from 100 mg of fresh leaves using a commercial kit (ExgeneTM; GeneAll Biotechnology Co. Ltd., Seoul, Korea). A total of 4.36 Gbp of paired-end reads is produced based on the MGISEQ platform with the MGIEasy FS DNA library kit (insert size: 500 bp) (MGI Tech Co. Ltd., Shenzhen, China). We mapped each paired-end read against cp genome of V. raddeana (NC_041584), a sister group of V. verecunda in a previous study (Yoo and Jang Citation2010), using Geneious prime (v. 11.0.11 + 7; Kearse et al. Citation2012) with a ‘medium-low’ sensitivity option. After the manual editing, the consensus sequence was annotated using GeSeq (Tillich et al. Citation2017) with six reference sequences from Viola (NC_041585.1, NC_026986.1, NC_041584.1, NC_052919.1, NC_041583.1, and NC_041582.1).
The cp genome of V. arcuata (GenBank accession number: OM301625) has 157,870 bp in length (GC ratio: 36.3%) and is composed of four regions: 86,366 bp of a large single-copy (LSC) region, 17,298 bp of a small single-copy (SSC) region, and 27,103 bp of a pair of inverted repeats (IRs). The cp genome has 111 unique genes, including 77 protein-coding genes, 4 rRNA genes, and 30 tRNA genes. Compared to the previously reported cp genome of V. verecunda (MW586692), V. arcuata has 27 bp of longer cp genome (21 bp in LSC and six bp in SSC). Thirty-four different loci (12 SNPs and 22 indels) were confirmed between the two genomes. Importantly, two amino acids were added at the 3′ end of ndhl in V. arcuata because of the frameshift caused by one base deletion. There are three synonymous substitutions (ndhF, ccsA, and ndhI) and six nonsynonymous substitutions (matK, rpoC2, ndhF, ycf1, and two rpl2s on each IR region) in the coding regions. In non-coding regions, 19 polyN variations, two single nucleotide polymorphisms (SNPs), two indels, and one microsatellite repeat variation were recognized.
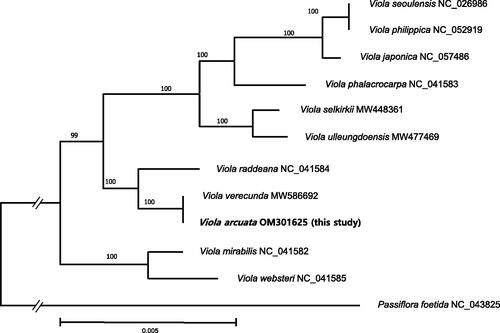
The phylogenetic tree () was produced with 11 complete cp genomes from Viola (including V. arcuata) and Passiflora foetida (Passifloraceae) as an outgroup (Xi et al. Citation2012). After aligning these sequences using MAFFT (v7.450; Katoh and Standley Citation2013), the maximum-likelihood analysis was performed using the MEGAX (Kumar et al. Citation2018) with a partial deletion (95%) option. The GTR + G model was selected as the best nucleotide substitution model by the Modeltest (Nei and Kumar Citation2000). The bootstrap was repeated 500 times to evaluate the reliability of each node with the same model and options. The result of the phylogenetic analysis confirms a sister or nearly identical relationship between two cp genomes from V. arcuata and V. verecunda.
Figure 1. A maximum-likelihood tree using 11 selected chloroplast genomes from Viola and an outgroup (Passifloraceae). Numbers above the node indicate bootstrap values.
This study reports the cp genome of V. arcuata collected in the Korean peninsula. Based on the present report, a comprehensive morphological and genomic study with various collections from East Asia will clarify the taxonomic identities of V. arcuata, V. verecunda, and V. hamiltoniana in the future.
Ethical approval
We disclose that V. arcuata, the material used in this study, is not an endangered species designated by the Ministry of Environment of the Republic of Korea and does not require permission for collection.
Author contributions
S. Kim designed the research, collected the material, produced raw data, and revised the manuscript. H. Moon analyzed data and prepared a preliminary manuscript. All authors have read and agreed to the published version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. OM301625. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA815741, SRR18318783, and SAMN26637850, respectively.
Additional information
Funding
References
- Akiyama S, Ohba H, Tabuchi S. 2002. Violaceae. In: Iwatsuki K, Boufford DE, Ohba H, editors. Flora of Japan c. Tokyo: Kodansha Ltd.
- Blume CL. 1825. Bijdragen tot de flora van Nederlandsch Indië 1. Vatavia: ter lands Drukkerij.
- Chang CS, Kim H, Chang KS. 2014. Provisional checklist of vascular plants for the Korean Peninsula Flora (KPF). Seoul: Designpost.
- Chayamarit K, Balslev H. 2018. Flora of Thailand 14. Bangkok: The Forest Herbarium, Royal Forest Department.
- Chen Y, Qiner Y, Ohba H, Nikitin VV. 2007. Viola L. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China 13. Beijing: Science Press.
- Cheon KS, Kim KA, Kwak M, Lee B, Yoo KO. 2019. The complete chloroplast genome sequences of four Viola species (Violaceae) and comparative analyses with its congeneric species. PLOS One. 14(3):e0214162.
- Don D, Wallich N, Vuchanan FH. 1825. Prodromus Florae Nepalensis. Londini: Veneunt apud J. Gale.
- Gray A. 1857. Memoirs of the American Academy of Arts and Sciences 6, Cambridge and Boston: Metcalf and company.
- Juzepchuk SV, Klokov MV. 1949. Violaceae. In Shishkin BK, Bobrov EG, editors. Flora of the U.S.S.R. 15. Moskva-Leningrad: Izdatel’stvo Akademii Nauk SSSR.
- Kang JI, Seo MJ, Choi YK, Shin SY, Hwang Y, Yoo ES, Kim SC, Kang HK. 2021. The mechanism of whole plant extract of Viola verecunda on the proliferation of dermal papilla cells. Korean J Pharmacogn. 52:34–40.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Kwak M. 2021. The complete chloroplast genome sequence of Viola verecunda (Violaceae). Mitochondrial DNA B Resour. 6(12):3409–3410.
- Lee WT, Yoo KO. 2020. Violaceae Batsch. In Park C, editor. Flora of Korea 4a. Incheon: National Institute of Biological Resources.
- Marcussen T, Ballard HE, Danihelka J, Flores AR, Nicola MV, Watson JM. 2022. A revised phylogenetic classification for Viola (Violaceae). BioRxiv doi: 10.1101/2022.04.22.489152.
- Moon HI, Jung JC, Lee J. 2007. Antiplasmodial activity of triterpenoid isolated from whole plants of Viola genus from South Korea. Parasitol Res. 100(3):641–644.
- Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. New York: Oxford university press.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wahlert GA, Marcussen T, De Paula-Souza J, Feng M, Ballard HE. 2014. A phylogeny of the Violaceae (Malpighiales) inferred from plastid DNA sequences: implications for genetic diversity and intrafamilial classification. Syst Bot. 39(1):239–252.
- Xi Z, Ruhfel BR, Schaefer H, Amorim AM, Sugumaran M, Wurdack KJ, Endress PK, Matthews ML, Stevens PF, Mathews S, et al. 2012. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc Natl Acad Sci USA. 109(43):17519–17524.
- Yoo KO, Jang SK. 2010. Infrageneric relationships of Korean Viola based on eight chloroplast markers. J Syst Evol. 48(6):474–481.
