Abstract
Persicaria maackiana (Regel) Nakai ex T. Mori (1922), a species of the Polygonaceae family, is an annual plant widely distributed in Northeast Asia. We aimed to sequence the complete chloroplast genome of P. maackiana using Illumina HiSeq paired-end sequencing. The chloroplast genome was determined to be 160,635 bp. The complete chloroplast genome contained 129 genes, including 84 protein-coding genes, 37 tRNA, and eight rRNA genes. Phylogenetic analysis of the chloroplast genome sequences of 15 Polygonaceae plants revealed that P. maackiana was most closely related to P. perfoliata. Our findings might be useful for future phylogenetic studies of Polygonaceae.
Persicaria maackiana (Regel) Nakai ex T. Mori is an annual plant of the Polygonaceae family, and the genus Persicaria. P. maackiana inhabits lowlands with fresh water, and is widely distributed in Northeast Asia – from southeastern China to Japan, Siberia, and the Korean Peninsula (Ohwi Citation1965; Komarov et al. Citation1968; Li et al. Citation2003; Chang et al. Citation2014). Numerous plant species of the Polygonaceae family have been used medicinally since ancient times (Wang et al. Citation1988; Hussain et al. Citation2010; Khatun et al. Citation2015; Tonny et al. Citation2017). Particularly, plants belonging to the genus Persicaria have often been used as diuretics and anti-inflammatory as well as to treat skin diseases, such as ringworm and boils (Khatun et al. Citation2015; Tonny et al. Citation2017). Recently, P. maackiana extract has been studied for its antidiabetic effect by promoting glucose absorption in human cells, and has been identified as a potential medicinal plant in Korea (NNIBR Citation2021). Despite its potential economic value, molecular genetic studies of P. maackiana have not been conducted in Korea. Therefore, we sequenced the complete chloroplast genome of P. maackiana as a first step to elucidate its genetic characteristics.
The P. maackiana leaf samples used in this study were collected from the Gaecheon Reservoir (36°23′42″ N, 128°27′56″ E) in Uiseong-gun, Gyeongsangbuk-do, South Korea (storage: Nakdonggang National Institute of Biological Resources; voucher number: NNIBRVP70284, email: [email protected]). High-quality genomic DNA was extracted using the DNeasy® Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Genomic DNA was sequenced using Illumina HiSeq sequencing with a 150 bp paired-end library. Moreover, 145 Gb of raw reads were obtained using Illumina HiSeq 2500 sequencing. We used NOVOPlasty v.4.3.1 (Dierckxsens et al. Citation2017) to assemble the complete chloroplast genome and CPGAVAS2 (Shi et al. Citation2019) to annotate the genome (Kearse et al. Citation2012). In addition, erroneous annotations were checked using National Center for Biotechnology Information (NCBI) BLAST and manually corrected using Geneious 11.0.12 software. The MISA tool (http://pgrc.ipk-gatersleben.de/misa/misa.html) was used to identify simple sequence repeat (SSR) regions in the chloroplast genome. The annotated chloroplast genome of P. maackiana was deposited into GenBank with the accession number OM386813.
The length of the complete chloroplast genome of P. maackiana was 160,635 bp, and the GC content was 37.9%. The GC content of the chloroplast genome of the genus Persicaria was 37.8%, 38.0%, and 38.0% in P. filiformis, P. chinensis, and P. perfoliata, respectively. Notably, the GC content of P. perfoliata chloroplast genome was most similar to that of P. maackiana.
The chloroplast genome of P. maackiana had a quadrilateral structure with a large single-copy (LSC) region of 85,375 bp, a short single-copy (SSC) region of 13,095 bp, and two inverted repeat (IR) regions of 31,131 bp. Furthermore, 129 functional genes were encoded, which included 84 protein-coding, 37 tRNA, and eight rRNA genes. We identified 51 SSRs, including 32 mononucleotides, six di-nucleotides, five tri- and tetra-nucleotides, two penta-nucleotides, and one hexa-nucleotide (S1 Table). Most SSRs were identified in the LSC and SSC regions (41), and the remaining in IR region a (5) and IR region b (5).
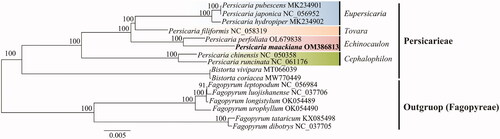
To determine the phylogenetic location of P. maackiana, chloroplast genome sequence of 15 Polygonaceae species was aligned using the MAFFT v.7.490 automated algorithm (Katoh and Standley Citation2013). The optimal GTR + G + I model was applied according to the Akaike information criterion using jModelTest v.2.1.7 to obtain the optimal model sequence (Posada Citation2008). The phylogenetic tree was reconstructed using the PhyML 3.0 program as the maximum-likelihood (ML) method, and 100 bootstrap replicates were performed (Guindon et al. Citation2010). Phylogenetic trees were visualized and manually edited using Figtree v1.4.4 (Rambaut Citation2018).
The phylogenetic tree was divided into two clades, Persicarieae and Fagopyreae tribes, with Fagopyreae as an external group. The first clade, Persicarieae, was clustered with the species of the genera Persicaria and Bistorta, and the species of the genus Persicaria formed a single clade with very high bootstrap values of over 99% (). The phylogenetic results of the ML analysis revealed that P. maackiana was most closely related to P. perfoliata. Notably, previous classical classification and phylogenetic results indicate that P. maackiana clusters with species of the section Echinocaulon (Park Citation1986; Kim and Donoghue 2008; Schuster et al. Citation2015). In this study, P. maackiana was clustered with P. perfoliata of the section Echinocaulon, which is consistent with previous studies (Park Citation1986; Kim and Donoghue 2008; Schuster et al. Citation2015). The P. maackiana chloroplast genome we sequenced might provide a solid basis for future genome-based phylogenetic and evolutionary relationship studies of Polygonaceae.
Ethical approval
No permission from the Republic of Korea government was required to collect these plants.
Consent form
The authors complied with relevant institutional (Nakdonggang National Institute of Biological Resources), national (Republic of Korea), and international guidelines (IUCN) and legislation for the plant study.
Author contributions
Kang-Rae Kim: conceptualization and data curation, NGS data analysis, writing – original draft, writing – review, and editing. So Young Park: NGS data analysis and data curation. Sun-Yu Kim: conceptualization, sampling, and investigation. Young Taek Oh: conceptualization, sampling, and investigation. Jeong-Nam Yu: conceptualization, data curation, supervision, funding acquisition, project administration, writing – review, and editing.
Supplemental Material
Download MS Excel (11.4 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession number OM386813. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA826290, SRR18740272, and SAMN27554296, respectively.
Additional information
Funding
References
- Chang CS, Kim H, Chang KS. 2014. Provisional checklist of vascular plants for the Korea Peninsula Flora (KPF). Seoul, Korea: Designpost.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Hussain F, Ahmad B, Hameed I, Dastagir G, Sanaullah P, Azam S. 2010. Antibacterial, antifungal and insecticidal activities of some selected medicinal plants of Polygonaceae. Afr J Biotechnol. 9(31):5032–5036.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:10.1093/bioinformatics/bts199.
- Khatun A, Imam MZ, Rana MS. 2015. Antinociceptive effect of methanol extract of leaves of Persicaria hydropiper in mice. BMC Complement Altern Med. 15(1):63.
- Kim ST, Donoghue MJ. 2008. Molecular phylogeny of Persicaria (Persicarieae, Polygonaceae). Syst Bot. 33(1):77–86.
- Komarov VL, Ryu R, Shishkin BK. 1968. Flora of the USSR. Vol. IV. Jerusalem: Israel Program for Scientific Translation Ltd.
- Li AJ, Bao BJ, Grabovskaya-Borodina AE, Hong SP, McNeill JM, Mosyakin SL, Hba HO, Park CW. 2003. Flora of China. Vol. 5 (Polygonaceae). Beijing; St. Louis: Science Press; Missouri Botanical Garden Press.
- NNIBR. 2021. Development of functional biomaterials using natural extract derived from freshwater bioresources. Sangju, Republic of Korea: Nakdonggang National Institute of Biological Resources; p. 1–26.
- Ohwi G. 1965. Flora of Japan [English translation]. Washington (DC): Smithsonian Institution.
- Park CW. 1986. Nomenclatural typifications in Polygonum section Echinocaulon (Polygonaceae). Brittonia. 38(4):394–406.
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7):1253–1256.
- Rambaut A. 2018. FigTree v1.4.4. Edinburgh: Institute of Evolutionary Biology University of Edinburgh; [accessed 2022 Feb 15]. http://tree.bio.ed.ac.uk/software/figtree/.
- Schuster TM, Reveal JL, Bayly MJ, Kron KA. 2015. An updated molecular phylogeny of Polygonoideae (Polygonaceae): relationships of Oxygonum, Pteroxygonum, and Rumex, and a new circumscription of Koenigia. Taxon. 64(6):1188–1208.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Tonny TS, Sultana S, Siddika F. 2017. Study on medicinal uses of Persicaria and Rumex species of Polygonaceae family. J Pharmacogn Phytochem. 6(6):587–589.
- Wang W, Wang JH, Shi TR. 1988. Effect of Polygonum multiflorum on the life-span and lipid metabolism in senile Japanese quails. Zhong Xi Yi Jie He Za Zhi. 8(4):223–224.