Abstract
African otter shrews (Potamogalidae) are Afrotherian mammals adapted to a semi-aquatic lifestyle. Given their rareness, genetic data on otter shrews are limited. By applying laboratory methods tuned for the recovery of archival DNA and an iterative mapping approach, we reconstructed whole mitochondrial genomes of the Giant (Potamogale velox) and Ruwenzori pygmy otter shrew (Micropotamogale ruwenzorii) from historical museum skins. Phylogenetic analyses are consistent with previous reports in recovering a sister relationship between African otter shrews and Malagasy tenrecs. The long branches separating both lineages, however, support their recognition as separate families.
Keywords:
Otter shrews (Potamogalidae) are neither otters nor shrews but most closely related to Malagasy tenrecs (Tenrecidae). Together with golden moles (Chrysochloridae), sengis (Macroscelididae) and the aardvark (Tubulidenta), they form the Afroinsectiphilia as the sister group to herbivorous Paenungulata (elephants, sea cows, hyraxes) within endemic African mammals (Afrotheria) (van Dijk et al. Citation2001). With only three species in two genera left (Giant and Pygmy otter shrews, Potamogale and Micropotamogale, respectively), otter shrews represent the remnants of an ancient radiation of African insectivores. They inhabit tropical river systems of central and western Africa and have a semi-aquatic lifestyle that includes foraging on fish, crabs, and other aquatic animals, and resting in burrows in the bankside (Vogel Citation2013). They exhibit striking phenotypic resemblance to small, semi-aquatic carnivores such as otters, minks, and fishers in having a streamlined body shape, a dense and water-repellent fur, closable nares, reduced olfaction, small eyes, and a flattened tail (Vogel Citation2013). Due to poaching, forest fragmentation and habitat destruction, otter shrew populations are declining, and two species are already listed as Endangered or Near Threatened by the IUCN. Their remote lifestyle, low abundance and low species number prevented intensive research on otter shrews so far, making them one of the least-studied mammalian families. Accordingly, genetic resources on otter shrews are very limited and complete mitochondrial genomes have not yet been published. In the absence of available fresh tissue samples, we assembled complete mitogenomes for representatives of both genera from historical samples by applying techniques tuned for the recovery of archival DNA.
The samples used in this study were collected from historical skins housed at the Royal Museum for Central Africa in Tervuren, Belgium (contact: [email protected]). The female Potamogale velox (du Chaillu, 1860) specimen (voucher No.: RMCA33978) was collected near Bionga, D. R. Congo (3.21°S, 28.08°E) on 19.11.1965. The male Micropotamogale ruwenzorii (de Witte and Frenchkop, 1955) specimen (voucher No.: RMCA31435) was collected near Butembo, D. R. Congo (0.09°N, 29.17°E) on 06.01.1963. A small piece of skin was sampled from the edge of each museum specimen (i.e. skin from the ventral trunk). DNA was extracted in the dedicated archival DNA facilities at the University of Potsdam using the protocol of Dabney et al. (Citation2013) with a nondestructive extraction buffer (Rohland et al. Citation2004). DNA extracts were built into single-stranded libraries following Gansauge et al. (Citation2017). The optimal number of amplification cycles for the subsequent dual-indexing PCR was determined with qPCR. Each library was shotgun sequenced using custom primers on the Illumina NextSeq500 system at the University of Potsdam, generating 20 million 75 bp single-end reads. Raw reads were trimmed with cutadapt v3.4 (Martin Citation2011).
Using an iterative mapping approach in MITObim v1.9.1 (Hahn et al. Citation2013) and the Echinops telfairi sequence (NC_002631.2) as reference bait, we obtained whole mitogenome sequences of 16,380 and 16,447bp for P. volex and M. ruwenzorii, respectively (but each including a short gap of 102 and 50 N’s, respectively). The mitogenomes were annotated with the MitoFinder pipeline (Allio et al. Citation2020) and publicly deposited (GenBank Accession No. OM912806 for P. volex and OM912807 for M. ruwenzorii). Protein-coding and rRNA genes were aligned with respective sequences of 14 other representatives of Afroinsectiphilia (four Malagasy tenrecs, two golden moles, seven sengis, one aardvark) in Mafft v7.490 (Katoh and Standley Citation2013) and subsequently subjected to a maximum likelihood phylogenetic analysis with IQ-TREE v2.14 (Nguyen et al. Citation2015; Hoang et al. Citation2018). The appropriate partition scheme and substitution models were determined with ModelFinder (Kalyaanamoorthy et al. Citation2017).
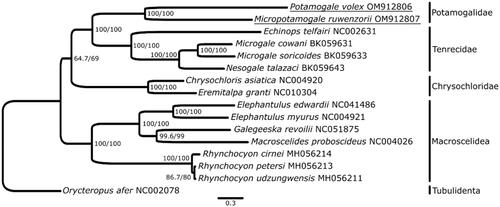
The phylogenetic analysis is consistent with previous studies in placing otter shrews as sister-clade to Malagasy tenrecs (). The long branches between them support their recognition as separate families (Everson et al. Citation2016). Furthermore, the long terminal branches of both otter shrew species suggest a deep divergence and long independent evolution of both lineages (Vogel Citation2013). All nodes of the tree are well supported except the one defining the sister-clade relationship of golden moles and the otter shrew plus Malagasy tenrec clade. This finding is in accordance with previously reported difficulties in establishing the phylogenetic relationships among afroinsectiphilian orders using genetic and genomic data (reviewed in Murphy et al. Citation2021). Thus, whole-genome data are still needed in order to compare different markers to resolve the deep divergences among placental mammal lineages (Lv et al. Citation2021).
Figure 1. Mitogenome phylogeny of selected Afroinsectiphilia. Support values are depicted next to the nodes (approximate likelihood ratio test for single branches/ultra-fast bootstrap).f
Overall, our newly assembled otter shrew mitogenomes further increase the (still limited) number of available mitogenomes of afroinsectiphilian Afrotheria as mitogenomic resources are available for all families now. In addition, our results highlight the value of historical museum samples for genetic research in rare species.
Ethical approval
No ethical approval or other relevant permission were needed for the study due to the use of historical museum material.
Author contributions
PA, EM and MH designed the study. PA and EG collected the tissue samples. PA and JH analyzed the data. PA and MH interpreted the data. PA drafted the manuscript. All authors critically revised the manuscript for intellectual content, approved the version to be published and agree to be accountable for all aspects of the work.
Acknowledgment
No funding was received.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The mitochondrial sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession numbers OM912806-OM912807. All project-related biological data are collected under BioProject number PRJNA836394. The associated BioSample and SRA numbers are SAMN28159688- SAMN28159689 and SRX15389535-SRX15389536, respectively.
References
- Allio R, Schomaker‐Bastos A, Romiguier J, Prosdocimi F, Nabholz B, Delsuc F. 2020. MitoFinder: efficient automated large‐scale extraction of mitogenomic data in target enrichment phylogenomics. Mol Ecol Resour. 20(4):892–905.
- Dabney J, Knapp M, Glocke I, Gansauge M-T, Weihmann A, Nickel B, Valdiosera C, García N, Pääbo S, Arsuaga J-L. 2013. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci USA. 110(39):15758–15763.
- Everson KM, Soarimalala V, Goodman SM, Olson LE. 2016. Multiple loci and complete taxonomic sampling resolve the phylogeny and biogeographic history of tenrecs (Mammalia: Tenrecidae) and reveal higher speciation rates in Madagascar’s humid forests. Syst Biol. 65(5):890–909.
- Gansauge M-T, Gerber T, Glocke I, Korlevic P, Lippik L, Nagel S, Riehl LM, Schmidt A, Meyer M. 2017. Single-stranded DNA library preparation from highly degraded DNA using T4 DNA ligase. Nucleic Acids Res. 45(10):e79–e79.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Research. 41(13):e129.
- Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lv X, Hu J, Hu Y, Li Y, Xu D, Ryder OA, Irwin DM, Yu L. 2021. Diverse phylogenomic datasets uncover a concordant scenario of laurasiatherian interordinal relationships. Mol Phylogenet Evol. 157:107065.
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10–12.
- Murphy WJ, Foley NM, Bredemeyer KR, Gatesy J, Springer MS. 2021. Phylogenomics and the genetic architecture of the placental mammal radiation. Annu Rev Anim Biosci. 9:29–53.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Rohland N, Siedel H, Hofreiter M. 2004. Nondestructive DNA extraction method for mitochondrial DNA analyses of museum specimens. Biotechniques. 36(5):814–821.
- van Dijk MA, Madsen O, Catzeflis F, Stanhope MJ, de Jong WW, Pagel M. 2001. Protein sequence signatures support the African clade of mammals. Proc Natl Acad Sci USA. 98(1):188–193.
- Vogel P. 2013. Subfamily Potamogalinae - Otter Shrews. In: Kingdon J, Happold D, Hoffmann M, Butynski T, Happold M, Kalina J, editors. Mammals of Africa volume I. Introductory chapters and Afrotheria. London: Bloomsbury; p. 216–222.
