Abstract
The Pygmy corydoras Corydoras pygmaeus Knaack, 1966, is the smallest member of the genus Corydoras, belonging to the family Callichthyidae and order Siluriformes. The complete mitochondrial genome of C. pygmaeus was sequenced and assembled using next-generation sequencing technology, and phylogenetically compared with those of other species of this genus. The mitochondrial genome of C. pygmaeus is a circular DNA molecule with a size of 16,840 bp (GenBank no. ON729306). A phylogenetic tree was constructed based on 13 protein-coding genes of C. pygmaeus and 13 species of the family Callichthyidae, which showed that C. pygmaeus clustered with other species of this genus, but was the first branch to differentiate. These results could provide basic data for phylogenetic analysis and population genetic diversity protection of Corydoras and Callichthyidae fish in the future.
Introduction
Compared to nuclear genes, mitochondrial DNA (mtDNA) is maternally inherited, undergoes rapid evolution, and small in size, making it an important molecular marker for research on fish evolutionary genetics, molecular ecology, species identification, and conservation biology (Sun et al. Citation2020). Mouse fish are members of the genera Aspidoras, Brochis, and Corydoras of the subfamily Corydoradinae. They have two cute ‘moustaches’ next to their mouths and look like small mice swimming in water, and thus the name ‘mouse fish.’ Mouse fish are typical benthic and migratory fish with omnivorous feeding habits. They are found in South America. Almost all the main and tributary water systems of the Amazon River contain pikes, but the composition of species in the watersheds where various species are distributed is different (Huysentruyt and Adriaens Citation2005; Liu et al. Citation2019; Lv et al. Citation2020; Tencatt et al. Citation2021; Sun et al. Citation2022). For instance, most mouse fish gather in the middle and lower reaches, where the water flow is relatively gentle, and a few strong swift warriors live in the upper reaches of the river. The Pygmy corydoras Corydoras pygmaeus () Knaack, 1966, is the smallest member of the genus Corydoras.
Materials
The sample was collected from Duyun City, Guizhou, China (26°17′31.15″ N, 107°31′13.02″ E) on June 10, 2021. This study was reviewed and approved by the Animal Ethics Committee of the Qiannan Normal University for Nationalities. Voucher specimens were stored at the School of Tourism and Resource Environment, Qiannan Normal University for Nationalities, Guizhou, China, under the voucher number C_pygmaeus_A (https://www.sgmtu.edu.cn/, Huajun Zhang, [email protected]).
Methods
DNA extraction was performed using a column animal genome DNA extraction kit according to the manufacturer’s protocol (Sun et al. Citation2021). Sequencing of the mitochondrial genome was performed on an Illumina Novaseq 6000 platform and assembled using GetOrganelle 1.7.5 (Jin et al. Citation2020). Genes in the sequence were annotated using MitoAnnotator (Iwasaki et al. Citation2013) (http://mitofish.aori.u-tokyo.ac.jp/annotation/input.html) and MITOS (http://mitos.bioinf.uni-leipzig.de/index.py). The tRNAscan-SE 1.21 (Lowe and Eddy Citation1997) (http://lowelab.ucsc.edu/tRNAscan-SE/index.html) online tool was used to predict the secondary structure of tRNA, which was corrected manually and finally submitted to GenBank (Accession no. ON729306). To determine the phylogenetic significance of C. pygmaeus in the genus Corydoras, 11 other species of this genus were used to reconstruct the phylogenetic tree based on 13 protein-coding genes, using Hemigrammus erythrozonus as the outgroup (Sun et al. Citation2021). The phylogenetic tree was constructed using the maximum likelihood method with IQ-TREE v1.6.8 under the edge-linked partition model for 5,000 standard bootstraps.
Results
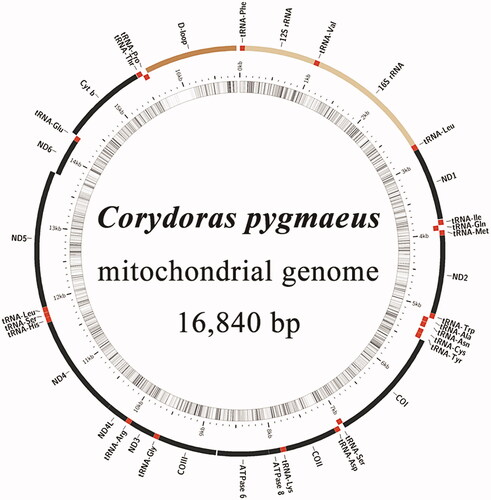
The complete mitochondrial genome of C. pygmaeus is 16,840 bp in length (), with a nucleotide composition of 33.25% A, 25.37% C, 14.39% G, and 26.98% T, and a higher A + T (60.24%) than G + C (39.76%) content. The mitochondrial genome is composed of a control region, 2 rRNA genes, 13 protein-coding genes, and 22 tRNA genes. Among these 37 genes, 9 are encoded by the light strand, while 28 are encoded by the heavy strand. All 13 protein-coding genes start with the codon ATG, except for COI, which starts with the codon GTG. Typical stop codons (TAG and TAA) were observed for most protein-coding genes, except for incomplete terminal codons T for ND2, COII, COIII, ND3, and ND4; AGA for ND5; and AGG for COI.
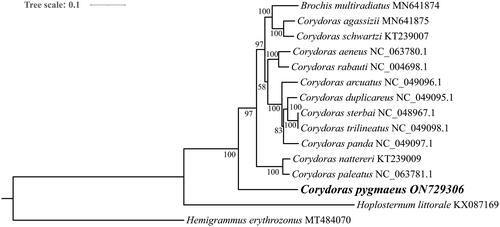
Phylogenetic analysis showed that Callichthyidae species formed a branch and were well separated from the outgroup, and that Brochis multiradiatus clustered with other species of the genus Corydoras before C. pygmaeus (). The target species C. pygmaeus clustered with other species of this genus, but was the first branch to differentiate.
Discussion and conclusion
In this study, the composition and structure of C. pygmaeus mtDNA were predicted and analyzed to provide basic data for phylogenetic analysis and population genetic diversity protection of Corydoras and Callichthyidae fish in the future.
Author contributions
Huajun Zhang: Conceptualization, Funding Acquisition, Investigation, Data Curation, Formal analysis, Writing – Original Draft; Li-An Gao: Formal analysis, Writing – Review & Editing; Wenlei Zhang: Formal analysis, Writing – Review & Editing; All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. ON729306. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA847725, SRR19612627, and SAMN28952161, respectively.
Additional information
Funding
References
- Huysentruyt F, Adriaens D. 2005. Adhesive structures in the eggs of Corydoras aeneus (Gill, 1858; Callichthyidae). J Fish Biol. 66(3):871–876.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):31.
- Liu Q, Xu B, Xiao T. 2019. Complete mitochondrial genome of Corydoras duplicareus (Teleostei, Siluriformes, Callichthyidae). Mitochondrial DNA B. 4(1):1832–1833.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Lv L, Su H, Xu B, Liu Q, Xiao T. 2020. Complete mitochondrial genome of Corydoras agassizii. Mitochondrial DNA B Resour. 5(1):727–728.
- Sun C-H, Huang Q, Zeng X-S, Li S, Zhang X-L, Zhang Y-N, Liao J, Lu C-H, Han B-P, Zhang Q. 2022. Comparative analysis of the mitogenomes of two Corydoras (Siluriformes, Loricarioidei) with nine known Corydoras, and a phylogenetic analysis of Loricarioidei. ZooKeys. 1083:89–107.
- Sun CH, Liu HY, Lu CH. 2020. Five new mitogenomes of Phylloscopus (Passeriformes, Phylloscopidae): sequence, structure, and phylogenetic analyses. Int J Biol Macromol. 146:638–647.
- Sun CH, Liu HY, Xu N, Zhang XL, Zhang Q, Han BP. 2021. Mitochondrial genome structures and phylogenetic analyses of two tropical characidae fishes. Front Genet. 12:627402.
- Tencatt LFC, Santos SA, Evers H‐G, Britto MR. 2021. Corydoras fulleri (Siluriformes: Callichthyidae), a new catfish species from the Rio Madeira basin, Peru. J Fish Biol. 99(2):614–628.