Abstract
Litsea honghoensis Liou is an endangered tree endemic to south China. In this study, the first complete plastid genome of L. honghoensis was presented, which had a length of 152,605 base pairs (bp) with a GC content of 39.20%. The genome consisted of a large single-copy (LSC) region of 93,560 bp, a small single-copy (SSC) region of 18,905 bp, and two inverted repeat regions (IRa and IRb) of 20,070 bp. There were 125 genes in the plastid genome, including 81 protein-coding genes, 36 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. Phylogenomic analysis based on 52 complete plastomes of Laureae in the family Lauraceae supports the close relationships among L. honghoensis, Lindera communis, Lindera nacusua, Lindera angustifolia, and Lindera glauca.
Keywords:
Litsea honghoensis Liou 1933 as an evergreen species of the family Lauraceae that is naturally distributed in the valley forests in the southeast and south of Yunnan, China (http://foc.iplant.cn/). It belongs to the genus Litsea Lamarck distributed mainly in tropical or subtropical Asia and North or South America (http://www.plantsoftheworldonline.org/). It is a key protected rare and vulnerable (VU) species in China (Qin et al. Citation2017), because of habitat degradation or loss, direct excavation or cutting, intrinsic factors and interspecific influence. However, there has been no genomic study on the L. honghoensis, which greatly limit the protection and utilization of L. honghoensis. Litsea is often confused with genus Lindera and Laurus in the core Lauraceae (Zhao et al. Citation2018; Tian et al. Citation2019; Liu et al. Citation2020; Song et al. Citation2020). In order to understand the relationships of L. honghoensis and other Laureae species, we assembled and annotated the plastid genome of L. honghoensis as a resource for evolution and protection research.
The leaf samples of L. honghoensis were collected from Kunming Arboretum (Kunming, China; Long. 102.740333 E, Lat. 25.134722 N, 1950 m). The voucher specimens (accession number: XTBG-BRG-SY35037) were deposited at the Herbarium of Xishuangbanna Tropical Botanical Garden. Total genomic DNA was extracted using the CTAB method (Doyle and Dickson Citation1987). A paired-end library of 350 bp was constructed. Approximately, 1.8 Gb of raw data of 150-bp-long paired-end reads were generated using the Illumina NovaSeq platform. The plastid genome of L. honghoensis was assembled and annotated using Geneious 4.8 and GeSeq (Tillich et al. Citation2017) with Lindera glauca (MG581443) served as the reference. The complete plastid genome of L. honghoensis was submitted to NCBI (accession number OL362095).
The whole plastid genome of L. honghoensis was 152,605 base pairs (bp) in length. The plastid genome exhibited a typical quadripartite structure with a large single-copy (LSC) region of 93,560 bp, a small single-copy (SSC) region of 18,905 bp, and two inverted repeat regions (IRs) of 20,070 bp. The plastid genome contained 125 genes, including 81 protein-coding genes, 36 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. The G + C content of the whole plastid genome was 39.20%, and those of LSC region, SSC region, and IR region were 38.02%, 33.93%, and 44.42%, respectively.
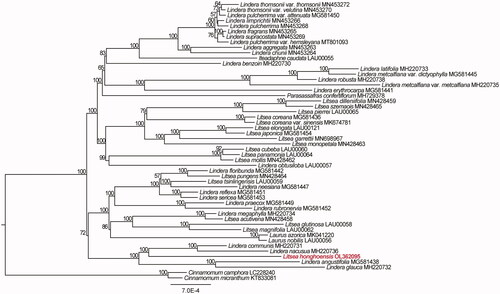
To further investigate the phylogenetic position of L. honghoensis, 52 complete plastid genome sequences in Laureae were aligned using MAFFT v. 7.490 (Katoh et al. Citation2019). Maximum-likelihood (ML) phylogenetic analyses were performed based on TIM + F+R2 model in the IQ-TREE v. 2.1.1 with 1000 bootstrap replicates (). The ML phylogenetic tree with 57–100% bootstrap values at each node showed that Litsea species was grouped into two clades, and that L. glutinosa, L. magnifolia, L. acutivena, L. honghoensis, L. pungens, and L. tsinlingensis were located in the same clade, while L. dilleniifolia, L. szemaois, L. coreana, L. coreana var. sinensis, L. elongata, L. garrettii, L. monopetala, L. mollis, L. cubeba, L. japonica, L. panamonja, and L. pierrei in another clade. L. honghoensis is closed related to Lindera communis, Lindera nacusua, Lindera angustifolia, and Lindera glauca with 100% bootstrap value. The Litsea is polyphyletic and closely related to the genera Lindera and Laurus, which is consistent with previous studies (Xiao et al. Citation2020; Zhang et al. Citation2021; Liu et al. Citation2022).
Ethics statement
The collection of specimen conformed to the requirement of International Ethics, which did not cause damage to the local environment. The process and purpose of this experimental research were in line with the rules and regulations of our institute. There are no ethical issues and other conflicts of interest in this study.
Author contributions
Lihong Han and Chao Liu were involved in the conception and design, analyses and interpretation of the data and writing of the manuscript; Jian Cai and Huanhuan Chen performed the analysis and interpretation of the data; Chao Liu revised it critically for intellectual content. All authors were involved in the final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Disclosure statement
We thanked Dr. Yu Song for providing and identifying the species of Litsea honghoensis. No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OL362095. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA778331, SRR16832243, and SAMN22942525, respectively.
Additional information
Funding
References
- Doyle JJ, Dickson EE. 1987. Preservation of plant samples for DNA restriction endonuclease analysis. Taxon. 36(4):715–722.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Liu C, Chen H, Han L, Tang L. 2020. The complete plastid genome of an evergreen tree Litsea elongata (Lauraceae: Laureae). Mitochondrial DNA B Resour. 5(3):2483–2484.
- Liu C, Chen HH, Tang LZ, Khine PK, Han LH, Song Y, Tan YH. 2022. Plastid genome evolution of a monophyletic group in the subtribe Lauriineae (Laureae, Lauraceae). Plant Divers. 44(4):377–388.
- Qin HN, Yang Y, Dong SY, He Q, Jia Y, Zhao LN, Yu SX, Liu HY, Liu B, Yan YH, et al. 2017. Threatened species list of China’s higher plants. Biodivers Sci. 25(7):696–744.
- Song Y, Yu WB, Tan YH, Jin JJ, Wang B, Yang JB, Liu B, Corlett RT. 2020. Plastid phylogenomics improve phylogenetic resolution in the Lauraceae. J Syst Evol. 58(4):423–439.
- Tian XY, Ye JW, Song Y. 2019. Plastome sequences help to improve the systematic position of trinerved Lindera species in the family Lauraceae. PeerJ. 7:e7662.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Xiao T, Xu Y, Jin L, Liu TJ, Yan HF, Ge XJ. 2020. Conflicting phylogenetic signals in plastomes of the tribe Laureae (Lauraceae). PeerJ. 8:e10155.
- Zhang Y, Tian Y, Tng DYP, Zhou J, Zhang Y, Wang Z, Li P, Wang Z. 2021. Comparative chloroplast genomics of Litsea Lam. (Lauraceae) and its phylogenetic implications. Forests. 12:744.
- Zhao ML, Song Y, Ni J, Yao X, Tan YH, Xu ZF. 2018. Comparative chloroplast genomics and phylogenetics of nine Lindera species (Lauraceae). Sci Rep. 8(1):8844.