Abstract
Stichopus ocellatus, known as eye-spotted sea cucumber, is a commercially important Stichopodidae holothuroid in Indo-Pacific region for its valuable nutrition and medicinal ingredients. However, because the taxonomic analyses based on morphological characters and molecular data within Aspidochirotida are limited, the deep-level evolutionary relationships of Aspidochirotida are still poorly understood. Here, for providing better insight of future evolutionary and taxonomic classification of Stichopodidae, we report the first complete mitogenome of S. ocellatus along with 37 annotated and characterized mitochondrial genes, and the phylogenetic analysis based on mitogenome data reveals sister relationship between S. ocellatus and S. monotuberculatus.
Stichopodidae, a family of sea cucumbers, is a diverse and conspicuous group of marine benthic ecosystems. Stichopodidae contains more than 128 living species, many of which are ecologically and economically important for enhancing ecosystem nutrient cycling and providing valuable nutrition (Uthicke et al. Citation2010). Despite being dominant large mobile invertebrates in local ecosystems, the taxonomy of many Stichopodidae species has been revised constantly due to the difficulty in application of traditional taxonomic characters for sea cucumbers (Byrne et al. Citation2010; Woo et al. Citation2015; Madduppa et al. Citation2017). The eye-spotted sea cucumber Stichopus ocellatus Massin, Zulfigar, Hwai & Boss, 2002, which inhabits the seabed in the Indo-Pacific region, was known as healthy seafood many years ago but was only described as a validated Stichopodidae species in 2002 (Massin et al. Citation2002). However, despite its commercial and ecological importance, the genetic diversity and taxonomy of S. ocellatus remain poorly explored due to insufficient molecular data and complex taxonomic characteristics, and the mitogenome of S. ocellatus has not been sequenced yet. The mitogenome data have proven to be an excellent molecular marker for studying genetic diversity and species identification (Uthicke et al. 2010; Madduppa et al. Citation2017). Here, we report the complete mitogenome sequence of S. ocellatus, which will provide a better understanding of genetic and taxonomic analyses in Stichopodidae.
Tissue samples of one individual S. ocellatus were collected from Hainan province, China (Dong Fang, 19.248723 N, 108.484287 E) by local diving fishermen and the whole body specimen (#JP0227) was deposited at Marine Biological Museum, Guangxi Institute of Oceanology, Beihai, China (http://www.gxas.cn/kypt/kxpj/kpcg, Shengping Zhong, [email protected]). The total genomic DNA was extracted from the muscle of the specimens using an SQ Tissue DNA Kit (OMEGA, Guangzhou, China) following the manufacturer’s protocol. DNA libraries (350 bp insert) were constructed with the TruSeq Nano™ kit (Illumina, San Diego, CA) and were sequenced (2 × 150 bp paired-end) using HiSeq platform at Novogene Company (Beijing, China). Mitogenome assembly was performed by MITObim (Hahn et al. Citation2013). The complete mitogenome of S. monotuberculatus (GenBank accession number: NC_052743) was chosen as the initial reference sequence for MITObim assembly. Gene annotation was performed by MITOS (Bernt et al. Citation2013).
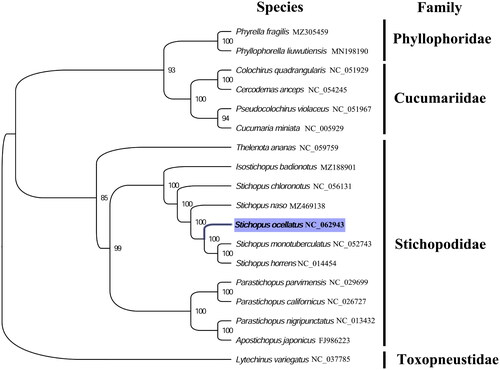
The complete mitogenome of S. ocellatus was 16,260 bp in length (GenBank accession number: NC_062943), and containing the typical set of 13 protein-coding, 22 tRNA, and two rRNA genes, and a putative control region. A total of 37 genes were annotated and 666 nucleotides were identified as putative control region. The overall base composition of the mitogenome was estimated to be A 31.6%, T 29.4%, C 23.8%, and G 15.6%, with a high A + T content of 60.6%, which is similar, but slightly higher than S. monotuberculatus (60.4%) within family Stichopodidae (Zhong et al. Citation2019). The phylogenetic analysis inferred from the concatenated nucleotides sequences of 13 PCGs shows that S. ocellatus clustered with S. monotuberculatus and S. horrens within family Stichopodidae (), which is consistent with the phylogenetic analyses of Stichopodidae holothuroids using mitochondrial COI and 16S sequences (Byrne et al. Citation2010; Madduppa et al. Citation2017). Our mitogenome data supported the sister relationship between S. ocellatus, S. monotuberculatus, and S. horrens. The complete mitogenome sequence of S. ocellatus adds one more mitogenome of family Stichopodidae, which will be useful for better resolving the genetic and taxonomic relationships of Stichopodidae, and related families.
Figure 1. Phylogenetic tree of 18 echinoderm species. The complete mitogenomes were downloaded from GenBank and the phylogenetic tree based on the concatenated nucleotide sequences of 13 mitochondrial PCGs was constructed by maximum-likelihood method via PhyML online server (http://www.atgc-montpellier.fr/phyml/), using GTR substitution model with 100 bootstrap replicates. The bootstrap values are indicated at each branch node, echinoid (Lytechinus variegatus) was chosen as the outgroup species.
Ethical approval
This study was conducted with the guidelines of the Council of China and animal welfare requirements. Based on the recommendations of the Regulations for the Administration of Affairs Concerning Experimental Animals of China, The Animal Care and Use Committee at Guangxi University of Chinese Medicine approved all animal experiments.
Author contributions
SZ and YL conceived and designed this study. LZ, GH, and LH performed bioinformatics analysis. SZ, LZ, and GH collected samples. SZ wrote the manuscript. YL edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. NC_062943. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA732759, SRR14654595, and SAMN19340382, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Byrne M, Rowe F, Uthicke S. 2010. Molecular taxonomy, phylogeny and evolution in the family Stichopodidae (Aspidochirotida: Holothuroidea) based on COI and 16S mitochondrial DNA. Mol Phylogenet Evol. 56(3):1068–1081.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.
- Madduppa H, Taurusman AA, Subhan B, Anggraini NP, Fadillah R, Tarman K. 2017. DNA barcoding reveals vulnerable and not evaluated species of sea cucumbers (Holothuroidea and Stichopodidae) from Kepulauan Seribu reefs. Biodiversitas. 18(3):893–898.
- Massin C, Zulfigar Y, Hwai TS, Boss SR. 2002. The genus Stichopus (Echinodermata: Holothuroidea) from the Johore Marine Park (Malaysia) with the description of two new species. Biologie. 72:73–99.
- Uthicke S, Byrne M, Conand C. 2010. Genetic barcoding of commercial Bêche-de-mer species (Echinodermata: Holothuroidea). Mol Ecol Resour. 10(4):634–646.
- Woo SP, Yasin Z, Tan SH, Kajihara H, Fujita T. 2015. Sea cucumbers of the genus Stichopus Brandt, 1835 (Holothuroidea, Stichopodidae) in Straits of Malacca with description of a new species. Zookeys. 545:1–26.
- Zhong S, Liu Y, Zhao Y, Huang G. 2019. The complete mitochondrial genome of sea cucumber Stichopus monotuberculatus (Aspidochirotida: Stichopodidae). Mitochondrial DNA B Resour. 4(2):3305–3306.
