Abstract
Priotyrannus closteroides Thomson, 1877 (Coleoptera: Cerambycidae) is the trunk borer of orange trees. In this study, we sequenced and annotated the whole mitochondrial genome of P. closteroides. The results showed that the length of the complete mitochondrial genome is 15,854 bp with an overall GC content of 32.11%. The genome encodes 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), and two ribosomal RNA genes (rRNAs). The relevant phylogenetic tree distinctly showed that P. closteroides is clustered with Dorysthenes paradoxus and Dorysthenes granulosus. This study provides a piece of valuable genomic information for the population genetics, evolution, and classification of P. closteroides.
Priotyrannus closteroides belongs to the insect subfamily Prioninae, within the Cerambycidae family which comprises more than 25,000 species (Sama et al. Citation2010). The bodies of P. closteroides adults are flat and wide, with russet feet and abdomen. The females are larger than males and the species prefers living in warm and humid environments. P. closteroides has been considered a trunk borer in forestry. In general, it attacks the orange trees roots which affects the growth and yield of the trees and causes plant death in the later stage. Therefore, it is important to study the biology and develop tools for control strategies. However, the information available about this species’ mitogenome and its taxonomic status is still limited. Here, we describe the complete mitochondrial genome of P. closteroides and the relevant phylogenetic tree. These results will support important genetic information for future studies related to the genetic evolution of P. closteroides.
The P. closteroides samples were obtained by using the sexual trapping method to trap in Hongwei, Fujian Province, China (118°57′12″E, 26°9′15″N). The voucher specimen (code TN-202103) was photographed and deposited at the Key Laboratory of Integrated Pest Management in Ecological Forests, Fujian Agriculture and Forestry University (contact person: Songqing Wu; email: [email protected]) (). Extraction and purification of the total genomic DNA was obtained by using the TruSeq DNA Sample Preparation Kit (Vazyme, Nanjing, China). The remaining part of the DNA was deposited in Dr. Wu’s lab with a unique code (DNA-PC1). A sequencing library with genomic DNA randomly broken into fragments using transposase was created and sequenced libraries with fragment peaks at 300 bp by using Agencourt SPRIselect Beads (Beckman Coulter, Indianapolis, IN) were selected by Shanghai Tianhao Biotechnology Company (Shanghai, China). To sequence the library preparations, we used the Illumina Hiseq 2500 (Illumina, San Diego, CA) with 2 × 150 bp paired-end reads (Yin et al. Citation2016). A total of 40,028,884 clean reads were assembled by using MitoZ and metaSPAdes (Nurk et al. Citation2017). Furthermore, the Mitos webserver was used to annotate the assembled sequences (Evans and Ślipiński Citation2016). To significantly improve the annotation obtained by Mitos was revised by using the closely related genome of Megopis sinica (Cerambycidae, GenBank accession number: MN594765) (Su and Wang Citation2020).
Figure 1. The phylogenetic tree and the picture of Priotyrannus closteroides Thomson, 1877. (A) Female of Priotyrannus closteroides Thomson, 1877, dorsal view. (B) Gene map of mitochondrial genome of Priotyrannus closteroides. (C) Maximum-likelihood tree of Priotyrannus closteroides Thomson and related 21 different insect species based on the complete mitochondrial genome. The numbers at each node represent bootstrap percentages of 1000 pseudoreplicates by ML analysis. The scale bar indicates the number of substitutions per site.
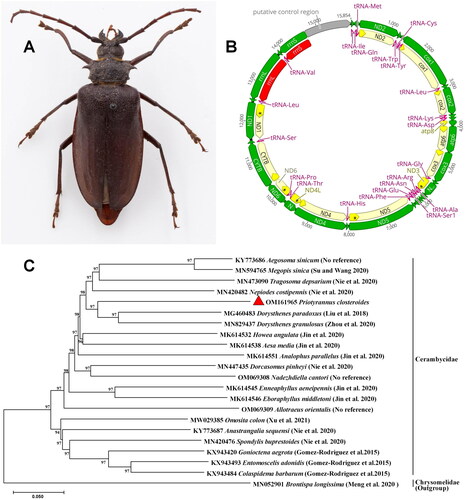
The results showed that the P. closteroides complete mitochondrial genome length is 15,854 bp, with 14,583 bp encoding open reading frames, with 13 protein-coding genes (PCGs) in Coleoptera typical order, 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs) (rrnS, rrnL) (Boore et al. Citation1998; Nie and Yang Citation2014). The mitochondrial genome map was visualized by Geneious Prime 2022.1.1 (). Among these 13 PCGs, three PCGs (atp6, nad3, nad4) start with ATA, four PCGs (nad2, atp8, nad5, nad1) start with ATT, two PCGs (cox2, nad6) start with ATC and three PCGs (cox3, nad4l, cob) start with ATG. However, the cox1 gene must use a non-conventional start site because no regular start codon is available after the last stop codon upstream to the cox1 open reading frame. The same phenomenon also has been found in other insects (De Oliveira et al. Citation2005; Castro et al. Citation2006; Lee et al. Citation2006; Sheffield et al. Citation2008). Also, 10 PCGs (nad2, cox1, cox2, atp6, cox3, nad5, nad4, nad4l, nad6, and cob) stop with TAA, three PCGs (atp8, nad3, and nad1) stop with TAG. The overall GC content is 32.11%. The annotated mitogenome sequence has been deposited in NCBI GenBank under accession number OM161965.
The mitochondrial sequences (including the nucleotide sequences of 13 PCGs) of P. closteroides and other mitochondrial genome of 20 Cerambycidae species selected from NCBI-BLAST (http://blast.ncbi.nlm.nih.gov) were used to construct phylogenetic tree with Brontispa longissima (Chrysomelidae; GenBank accession number: MN052901) (Meng et al. Citation2020) as an outgroup (Zhang et al. Citation2018). The 13 PCGs sequences were aligned using ClustalW (Thompson et al. Citation2002) in MEGA X (Kumar et al. Citation2018). Poorly aligned regions were excluded in Gblocks v.0.91b (Castresana Citation2000). PartitionFinder v2.1.1 (Lanfear et al. Citation2017) was run to select best-fit partitioning schemes and evolutionary models (GTR + G+I). We used RAxML v8.2.10 (Stamatakis Citation2014) based on the rapid bootstrap (BS) algorithm with 1000 BS replicates to construct maximum-likelihood (ML) tree. The phylogenetic analysis suggests that the mitochondrial genome of P. closteroides constitutes a monophyletic group with the mitochondrial genome of other 20 Cerambycidae species, and is in a close relationship with the mitochondrial genome of Dorysthenes paradoxus (Liu et al. Citation2018) and Dorysthenes granulosus (Zhou et al. Citation2020; ).
Ethics statement
This research does not involve ethical research. The collection of the specimens was carried out following the guidelines provided by the authors’ institution (Key Laboratory of Integrated Pest Management in Ecological Forests, Fujian Agriculture and Forestry University) and nation.
Author contributions
Xuanxuan Wu, Yiqi Lin, and Yunzhu Sun did the sample collection and laboratory work. Yiqi Lin photographed the species. Xuanxuan Wu, Songqing Wu, and Mingyong Luo were involved in the conception and design. Xuanxuan Wu, Yiqi Lin, Bowei Zhou, and Yunzhu Sun analyzed and interpreted the data. Xuanxuan Wu drafted the paper. Xuanxuan Wu revised the paper critically for intellectual content. All authors are involved: the final approval of the version to be published. All authors agreed to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data supporting this study’s findings are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under assessment no. OM161965. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA796508, SAMN24891047, and SRR17578369, respectively.
Additional information
Funding
References
- Boore JL, Lavrov DV, Brown WM. 1998. Gene translocation links insects and crustaceans. Nature. 392(6677):667–668.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552.
- Castro LR, Ruberu K, Dowton M. 2006. Mitochondrial genomes of Vanhornia eucnemidarum (Apocrita: Vanhorniidae) and Primeuchroeus spp. (Aculeata: Chrysididae): evidence of rearranged mitochondrial genomes within the Apocrita (Insecta: Hymenoptera). Genome. 49(7):752–766.
- De Oliveira MT, De Azeredo-Espin AML, Lessinger AC. 2005. Evolutionary and structural analysis of the cytochrome c oxidase subunit I (COI) gene from Haematobia irritans, Stomoxys calcitrans and Musca domestica (Diptera: Muscidae) mitochondrial DNA. DNA Seq. 16(2):156–160.
- Evans B, Ślipiński A. 2016. Review of the genus Tricheops Newman (Coleoptera: Cerambycidae: Cerambycinae) with description of two new species from Western Australia. Zootaxa. 4137(4):569–577.
- Gómez-Rodríguez C, Crampton-Platt A, Timmermans MJTN, Baselga AÉS, Vogler AP. 2015. Validating the power of mitochondrial metagenomics for community ecology and phylogenetics of complex assemblages. Methods Ecol Evol. 6(8):883–894.
- Jin M, Zwick A, Ślipiński A, Keyzer R, Pang H. 2020. Museomics reveals extensive cryptic diversity of Australian prionine longhorn beetles with implications for their classification and conservation. Syst Entomol. 45(4):745–770.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Lee ES, Shin KS, Kim MS, Park H, Cho SW, Kim CB. 2006. The mitochondrial genome of the smaller tea tortrix Adoxophyes honmai (Lepidoptera: Tortricidae). Gene. 373:52–57.
- Liu YQ, Chen DB, Liu HH, Hu HL, Bian HX, Zhang RS, Yang RS, Jiang XF, Shi SL. 2018. The complete mitochondrial genome of the longhorn beetle Dorysthenes paradoxus (Coleoptera: Cerambycidae: Prionini) and the implication for the phylogenetic relationships of the Cerambycidae species. J Insect Sci. 18(2):21.
- Meng R, Ao S, Xu W, Lv B, Cai B. 2020. Complete mitochondrial genome of coconut hispine beetle Brontispa longissima (Coleoptera: Chrysomelidae: Cassidinae). Mitochondrial DNA Part B. 5(1):1126–1127.
- Nie RE, Yang XK. 2014. Research progress in mitochondrial genomes of Coleoptera. Acta Entomol Sin. 57(7):860–868.
- Nie R, Vogler AP, Yang XK, Lin MY. 2020. Higher-level phylogeny of longhorn beetles (Coleoptera: Chrysomeloidea) inferred from mitochondrial genomes. Syst Entomol. 46(1):56–70.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Sama G, Buse J, Orbach E, Friedman A-L-L, Rittner O, Chikatunov V. 2010. A new catalogue of the Cerambycidae (Coleoptera) of Israel with notes on their distribution and host plants. Munis Entomol Zool. 5(1):1–51.
- Sheffield NC, Song H, Cameron L, Whiting MF. 2008. A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol Biol Evol. 25(11):2499–2509.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Su RR, Wang XY. 2020. The complete mitochondrial genome of the Megopis sinica white (Coleoptera: Cerambycidae: Prioninae). Mitochondrial DNA Part B. 5(1):236–237.
- Thompson JD, Gibson TJ, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics.
- Xu W, Wang Y, Wang M, Wang YH, Zhang YN, Wang JF. 2021. Characterization of the complete mitochondrial genome of Omosita colon (Coleoptera: Nitidulidae). Mitochondrial DNA Part B. 6(4):1547–1553.
- Yin D, Ritchie ME, Jabbari JS, Beck T, Blewitt ME, Keniry A. 2016. High concordance between Illumina HiSeq2500 and NextSeq500 for reduced representation bisulfite sequencing (RRBS). Genomics Data. 10(C):97–100.
- Zhang SQ, Che LH, Li Y, Liang D, Pang H, Slipinski A, Zhang P. 2018. Evolutionary history of Coleoptera revealed by extensive sampling of genes and species. Nat Commun. 9(1):205.
- Zhou SC, Zheng XL, Lu W. 2020. The complete mitochondrial genome of an Asian longicorn beetle Dorysthenes granulosus (Coleoptera: Cerambycidae: Prioninae). Mitochondrial DNA Part B. 5(1):673–674.
