Abstract
Rhododendron shanii W.P. Fang 1983 (Ericaceae) is woody plant naturally distributed in the southwest of Anhui, China. The complete chloroplast genome sequence of R. shanii was generated by whole-genome next-generation sequencing data and assembled based on three Rhododendron species chloroplast genome. The complete chloroplast genome sequence of R. shanii was 204,170 bp and divided into four distinct regions: small single-copy region (2615 bp), large single-copy region (107,189 bp), and a pair of inverted repeat regions (47,183 bp). The genome annotation displayed 150 genes, including 95 protein-coding genes, 47 tRNA genes, and eight rRNA genes. Phylogenetic analysis with the Ericaceae reported chloroplast genomes revealed that R. shanii is sister to the clade comprising R. delavayi, R. griersonianum and R. platypodum.
Rhododendron shanii Fang (Ericaceae) is a woody plant, which is mainly distributed in the mid-subtropical hilly areas of the southwest of Anhui province in China. R. shanii is a narrowly distributed species in the Dabie Mountain, its distribution range is only limited to a long and narrow zone with an area of 110 km2 in the junction district of Yuexi County, Huoshan County in Anhui Province and Yingshan County in Hubei Province, and mainly distributes on the ridge or on the slope near the ridge at altitude over 1400 m (Zhao et al. Citation2012). Zhao kai believes that according to their investigation results and classification standard of IUCN endangered species red list, it is determined that the endangered level of R. shanii should be vulnerable grade (VU) at least (Zhao et al. Citation2012). The plant is used as ornamental flowers, which is endemic to the southern Dabie Mountains (Zhao et al. Citation2010). Here, we reported the complete chloroplast (cp) genome of R. shanii, deposited the annotated cp genome into GenBank with the accession numbers MW374796.
Fresh leaves of R. shanii were collected from Duozhijian of Yuexi County (31°3′50″E 116°10′52″E), Anhui province, China. Then these leaves were dried with the silicone. Voucher specimens were deposited at Nanjing Institute of Environmental Sciences (http://ppbc.iplant.cn/tu/9352036, Huanxi Yu and [email protected]), and the voucher number of the specimen is DZJ202001. The genomic DNA was extracted following the modified CTAB method from the dry and healthy leaves (Doyle and Doyle Citation1987). The isolated genomic was manufactured to an average 400 bp paired-end (PE) library and sequenced by Illumina genome analyzer (Hiseq PE400). Sequencing service was provided by Personal Biotechnology Co., Ltd. Shanghai, China. The filtered reads were assembled using the program NOVOPlasty (Dierckxsens et al. Citation2017) with the complete cp genome of Rhododendron delavayi, R. griersonianum and R. platypodum as the reference (GenBank accession number MN413198, MT533181, MT985162 and MN711645). The assembled cp genome was annotated using Geneious 11.0.4 and corrected manually (Kearse et al., Citation2012). The complete cp genome of 11 species was aligned using MAFFT (Katoh et al. Citation2002). Total 11 species of Ericaceae were employed to build the maximum likelihood (ML) tree using RaxML (Stamatakis Citation2006) with 1000 bootstrap replicates. The cp genome of R. shanii was 204,170 bp in length (GenBank accession number MW374796), containing a large single-copy region (LSC) of 107,189 bp, a small single-copy region (SSC) of 2615 bp, and a pair of inverted repeat (IR) regions of 47,183 bp. Genome annotation predicted 150 genes, including 95 protein-coding genes, 47 tRNA genes, and 8 rRNA genes.
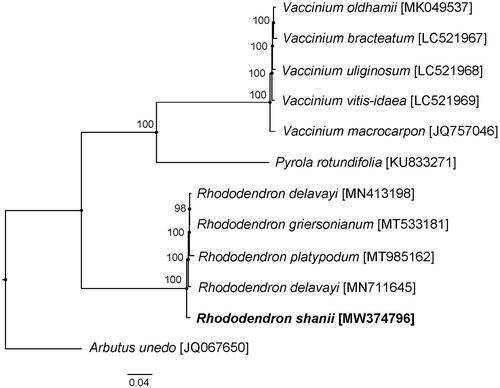
With released complete cp genome of Ericaceae, the phylogenetic analysis suggested that R. shanii is close to Rhododendron delavayi Franch., Rhododendron griersonianum Balf. f. & Forrest and Rhododendron platypodum Diels (). Arbutus unedo is the outgroup in the phylogenetic analysis. Our study represents the fist look into the complete cp genome of R. shanii, with 2001 bp longer than that of R. delavayi (Li et al. Citation2020), 2297 bp shorter than that of R. griersonianum (Liu et al. Citation2020) and 3123 bp longer than that of R. platypodum (Ma et al. Citation2021) separately. This complete cp genome can provide a genomic resource and contribute to constructing phylogenetic relationships and evolutionary studies among the subgenus Hymenanthes (Blume) K. Koch. and relative groups.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors. I confirm that all the research meets ethical guidelines and adheres to the legal requirements of the study country.
Author contribution
Huan-Xi Yu and Wang-Gu Xu contributed to the study concept and design; Zhi Wang, Jian-Liang Zhang and Peng Chen contributed to the acquisition of the data; Ying-Ying Lv, Bao-Kun Xu and Chang Qu analyzed and interpreted the data; and so on All authors read and approved the final manuscript.
Acknowledgments
The authors appreciate the opened raw cp genome data in NCBI. The authors thank Xian-Lin Guo for help with the technology. The authors thank Yong Chu, Jun Chu for their help with collecting the materials.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete chloroplast genome data supporting this study have been deposited in GenBank under the accession number MW374796 and is also openly available at https://www.ncbi.nlm.nih.gov/nuccore/MW374796.1/. The associated BioProject, SRA, and Bio-Sample number are PRJNA792687, SRR17380253, and SAMN24450521, respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li HE, Guo QQ, Li Q, Lan Y. 2020. Long-reads reveal that Rhododendron delavayi plastid genome contains extensive repeat sequences, and recombination exists among plastid genomes of photosynthetic Ericaceae. PeerJ. 8:e9048.
- Liu D, Fu CN, Yin LJ, Ma YP. 2020. Complete plastid genome of Rhododendrongriersonianum, a critically endangered plant with extremely small populations (PSESP) from southwest China. Mitochondrial DNA B Resour. 5(3):3, 3086–3087.
- Ma LH, Zhu HX, Wang CY, Li MY, Wang HY. 2021. The complete chloroplast genome of Rhododendronplatypodum (Ericaceae): an endemic and endangered species from China. Mitochondrial DNA B Resour. 6(1):196–197.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Zhao K, Shao JW, Wang G, Wang GX. 2012. Analyses of geographical distribution and population status of Rhododendron shanni. J Plant Resour Environ. 21(3):93–97.
- Zhao K, Yang WH, Chen WL, Shao JW. 2010. A revise of stamen number to an endangered plant: Rhododendron shanii Fang. Bull Bot Res. 30(6):664–667.