Abstract
Axarus fungorum (Albu, 1980) exhibits certain adaptations to different aquatic environments, appearing as an important evaluation element for freshwater quality monitoring. In this study, complete mitogenome of A. fungorum was provided for the first time to define the systematic and phylogenetic history of this taxon. The whole mitogenome is 15,696 bp long with high A + T content that consists of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and a noncoding control region. ML analysis showed support for monophyly of Chironominae and close relationship between A. fungorum and Chironomus generic genera.
Keywords:
Axarus fungorum (Albu, 1980) belongs to Chironominae, a subfamily under the Chironomidae, one of the most abundant invertebrate taxa in freshwater ecosystems with more than 6300 valid species. Chironomid larvae are considered as an excellent indicator for monitoring aquatic environment quality due to their wide distribution, high species diversity, large population, sensitivity, and adaptability (Ferrington Citation2008). Due to species diversity and variable morphological features within Chironomidae, the traditional morphological identification is inconvenient. In such instances, mitogenomic data can be considered as powerful and convenient material for molecular identification and phylogenetic studies for Diptera (e.g. Yan et al. Citation2019; Li et al. Citation2020; Zhang et al. Citation2022). However, complete mitogenomes are still scarce for Chironomidae (Beckenbach 2012; Kim et al. Citation2016; Deviatiiarov et al. Citation2017; Kong et al. Citation2021; Lei et al. Citation2021; Zheng et al. Citation2021, 2022; Fang et al. Citation2022). In the present study, we have provided complete mitochondrial genome of A. fungorum for the first time.
Fresh and adult male individuals of A. fungorum were collected from Meitan, Guizhou, China (27.828857°N, 107.5955098°E) on 8 June 2020. The DNeasy Blood and Tissue kit (QIAGEN Sciences, Valencia, CA) was used to isolate total genomic DNA from the muscle tissues of head and thorax. The DNA and voucher specimen of A. fungorum has been deposited in the College of Fisheries and Life Science, Shanghai Ocean University, Shanghai, China (https://www.shou.edu.cn, Xiao-Long Lin, [email protected]) under the voucher number DLC28. COI of A. fungorum (GenBank accession: MN521232) was used as bait to iterate and assemble the mitogenome of A. fungorum. DNA fragments with 350 bp insert size were sequenced by Illumina Nova6000 (PE150, Illumina, San Diego, CA) platform using pair-end strategy at Novogene Co., Ltd. (Cambridge, UK). Four Gb clean data were obtained from the library by trimming using Trimmomatic (Bolger et al. Citation2014). IDBA-1.1.1 (Peng et al. Citation2012) software package was employed to assemble the data. The bait sequence of COI (Crampton-Platt et al. Citation2015) was used in the BLAST program (Altschul et al. Citation1990) to compare with the mitogenome of A. fungorum. The percentage of match rate was found as 100% from the blast result. The mitogenome annotation was conducted as previously described by Zheng et al. (Citation2020).
The double-strand circular mitogenome of A. fungorum is 15,696 bp in length (GenBank accession no. ON099430) which encodes for 37 genes (13 protein-coding genes, two rRNA genes, and 22 tRNA genes) and a control region. Nucleotides within the mitogenome were distributed as follows: 41.2% A, 38.3% T, 12.2% C, and 8.3% G. The most frequently observed start codons were ATG for ATP6, COII, COIII, CytB, ND4, ND4L and ATT for ATP8, ND2, ND3, ND6, respectively, while GTG for ND5; TTG for COI and ND1. All of the 13 PCGs were terminated with TAA stop codon. Mitogenome organization, nucleotide composition and codon usage were similar to the previously sequenced Chironomidae mitogenomes with a high AT bias (79.5%).
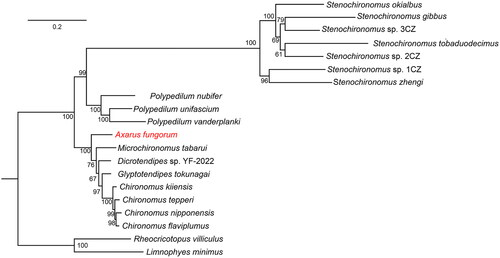
Eighteen mitogenomes of Chironominae and two of Orthocladiinae were mined from GenBank for the phylogenetic analysis. Initially, sequences of 13 PCGs were concatenated and then aligned with MAFFT (Katoh and Standley Citation2013) keeping all the settings in default (Katoh and Standley Citation2013). Using 1000 bootstraps and PMSF acid substitution model, we conducted phylogenetic analysis by maximum-likelihood (ML) method with IQ-TREE (Nguyen et al. Citation2015) considering Limnophyes minimus and Rheocricotopus villiculus as outgroups. Topologies from the reconstructed tree strongly supported the monophyly of Chironominae, and the sister relationship between A. fungorum and the Chironomus generic genera ().
Figure 1. Phylogenetic tree of 20 Chironomidae species based on the concatenated dataset of 13 PCGs using the maximum-likelihood (ML) method. The following sequences were used: Axarus fungorum ON099430 (present study), Chironomus flaviplumus MW770891 (Park et al. Citation2021), Chironomus kiiensis MZ150770 (Liu et al. Citation2022), Chironomus nipponensis MZ747092 (Shen et al. Citation2022), Chironomus tepperi JN861749 (Beckenbach Citation2012), Dicrotendipes sp. YF-2022 MZ747093 (direct submission), Glyptotendipes tokunagai MZ747091 (direct submission), Limnophyes minimus MZ041033 (Fang et al. Citation2022), Microchironomus tabarui MZ261913 (Kong et al. Citation2021), Rheocricotopus villiculus MW373526 (Zheng et al. Citation2021), Stenochironomus gibbus OL742440 (Zheng et al. Citation2022), Stenochironomus okialbus OL753645 (Zheng et al. Citation2022), Stenochironomus sp. 1CZ OL753646 (Zheng et al. Citation2022), Stenochironomus sp. 2CZ OL742441 (Zheng et al. Citation2022), Stenochironomus sp. 3CZ OL753647 (Zheng et al. Citation2022), Stenochironomus tobaduodecimus OL753648 (Zheng et al. Citation2022), Stenochironomus zhengi OL753649 (Zheng et al. Citation2022), Polypedilum nubifer MZ747090 (direct submission), Polypedilum unifascium MW677959 (Lei et al. Citation2021), and Polypedilum vanderplanki KT251040 (Deviatiiarov et al. Citation2017).
Ethics statement
The collection of specimen conformed to the requirement of International ethics, which did not cause damage to the local environment. The process and purpose of this experimental research were in line with the rules and regulations of our institute. There are no ethical issues and other conflicts of interest in this study.
Author contributions
Yan Qi and Xin Duan were involved in the conception and design, analysis, and interpretation of the data; Ke-Long Jiao and Xiao-Long Lin were involved in the drafting of the paper, revising it critically for intellectual content and the final approval of the version to be published; and the authors agreed to be accountable for all aspects of the work. No potential conflict of interest was reported by the authors.
Disclosure statement
No potential competing interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. ON099430. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA820975, PRJNA820975, and SAMN27029903, respectively.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410.
- Beckenbach AT. 2012. Mitochondrial genome sequences of Nematocera (lower Diptera): evidence of rearrangement following a complete genome duplication in a winter crane fly. Genome Biol Evol. 4(2):89–101.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Crampton-Platt A, Timmermans MJ, Gimmel ML, Kutty SN, Cockerill TD, Vun Khen C, Vogler AP. 2015. Soup to tree: the phylogeny of beetles inferred by mitochondrial metagenomics of a Bornean rainforest sample. Mol Biol Evol. 32(9):2302–2316.
- Deviatiiarov R, Kikawada T, Gusev O. 2017. The complete mitochondrial genome of an anhydrobiotic midge Polypedilum vanderplanki (Chironomidae, Diptera). Mitochondrial DNA A DNA Mapp Seq Anal. 28(2):218–220.
- Fang XL, Li X, Lu T, Fu J, Shen M, Xiao YL, Fu Y. 2022. Complete mitochondrial genome of Limnophyes minimus (Diptera: Chironomidae). Mitochondrial DNA Part B. 7(1):280–282.
- Ferrington LC. 2008. Global diversity of non-biting midges (Chironomidae; Insecta-Diptera) in freshwater. Hydrobiologia. 595(1):447–455.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim S, Kim H, Shin SC. 2016. Complete mitochondrial genome of the Antarctic midge Parochlus steinenii (Diptera: Chironomidae). Mitochondrial DNA Part A. 27(5):3475–3476.
- Kong FQ, Zhao YC, Chen JL, Lin XL. 2021. First report of the complete mitogenome of Microchironomus tabarui Sasa, 1987 (Diptera, Chironomidae) from Hebei Province, China. Mitochondrial DNA B Resour. 6(10):2845–2846.
- Lei T, Song C, Zhu XD, Xu BY, Qi X. 2021. The complete mitochondrial genome of a non-biting midge Polypedilum unifascium (Tokunaga, 1938) (Diptera: Chironomidae). Mitochondrial DNA Part B. 6(8):2212–2213.
- Li XY, Yan LP, Pape T, Gao YY, Zhang D. 2020. Evolutionary insights into bot flies (Insecta: Diptera: Oestridae) from comparative analysis of the mitochondrial genomes. Int J Biol Macromol. 149:371–380.
- Liu C, Xu G, Lei T, Qi X. 2022. The complete mitochondrial genome of a tropical midge Chironomus kiiensis Tokunaga, 1936 (Diptera: Chironomidae). Mitochondrial DNA Part B. 7(1):211–212.
- Nguyen LT, Schmidt HA, Haeseler AV, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Park K, Kim WS, Park JW, Kwak IS. 2021. Complete mitochondrial genome of Chironomus flaviplumus (Diptera: Chironomidae) collected in Korea. Mitochondrial DNA Part B. 6(10):2843–2844.
- Peng Y, Leung H, Yiu S, Chin F. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28(11):1420–1428.
- Shen M, Li Y, Yao Y, Fu Y. 2022. Complete mitochondrial genome of Chironomus nipponensis, new record from China (Diptera: Chironomidae). Mitochondrial DNA Part B. 7(6):1163–1164.
- Yan L, Pape T, Elgar MA, Gao Y, Zhang D. 2019. Evolutionary history of stomach bot flies in the light of mitogenomics. Syst Entomol. 44(4):797–809.
- Zhang X, Yang D, Kang Z. 2022. New data on the mitochondrial genome of Nematocera (lower Diptera): features, structures and phylogenetic implications. Zool J Linn Soc. zlac012.
- Zheng CG, Liu Z, Zhao YM, Wang Y, Bu WJ, Wang XH, Lin XL. 2022. First report on mitochondrial gene rearrangement in non-biting midges, revealing a synapomorphy in Stenochironomus Kieffer (Diptera: Chironomidae). Insects. 13(2):115.
- Zheng CG, Ye Z, Zhu XX, Zhang HG, Dong X, Chen P, Bu WJ. 2020. Integrative taxonomy uncovers hidden species diversity in the rheophilic genus Potamometra (Hemiptera: Gerridae). Zool Scr. 49(2):174–186.
- Zheng CG, Zhu XX, Yan LP, Yao Y, Bu WJ, Wang XH, Lin XL. 2021. First complete mitogenomes of Diamesinae, Orthocladiinae, Prodiamesinae, Tanypodinae (Diptera: Chironomidae) and their implication in phylogenetics. PeerJ. 9:e11294.
