Abstract
Although Mesogobio lachneri is the type species of the genus Mesogobio, its systematic position and status have remained unresolved to date. In this study, for the first time, we report the complete mitochondrial genome of M. lachneri using Sanger sequencing. It is a circular genome with a length of 16,602 bp, comprising 22 tRNAs, 13 protein-coding genes (PCGs), two rRNAs, and one non-coding control region. Our phylogenetic analysis reveals that M. lachneri is the close relative of the genus Gobio, indicating that Mesogobio may be a valid genus.
1. Introduction
The freshwater fish genus Mesogobio is placed into the order Cypriniformes, the family Gobionidae, and the sub-family Gobioninae (Tan and Armbruster Citation2018). The genus comprises two species Mesogobio lachneri Bănărescu & Nalbant 1973 and Mesogobio tumenensis Chang 1980 which are known to be distributed in Northeast Asia (Xie Citation2007). The results of two prior studies on the molecular phylogeny of the Gobioninae have indicated that M. tumensis should be moved into the Gobio, as G. tumensis (Yang et al. Citation2006; Tang et al. Citation2011). However, the type species (M. lachneri) of the genus Mesogobio has lacked molecular data until now. Hence, the systematic position and status of M. lachneri have remained unresolved. M. lachneri is a small benthic and rheophilic species with the body length less than 14 cm (). It prefers pebbly or sandy substrate environments, and is endemic to the Yalu River (the boundary river between North Korea and China) drainage (Yue Citation1998; Xie Citation2007). M. lachneri differs from other gobionid species, in having the following six morphological characters: a naked breast, absence of sub-lobes on the lower lip, absence of mental barbels, two rows of teeth, lips with developed papillae, and scale rows above lateral lines 5.5 (Yue Citation1998).
Figure 1. Female Mesogobio lachneri. The specimen from the Linjiang City, Jilin Province, China. The photograph by Cuizhang Fu on 22 September 2017.

In the present study, the complete mitochondrial genome of M. lachneri is obtained for the first time, which may be used to clarify the phylogenetic position of the genus Mesogobio within the family Gobionidae in the future. The mitogenome is also useful as a reference sequence for molecular species identification, as well as for further research on the molecular evolution of the family Gobionidae.
2. Materials
M. lachneri was collected from the Yalu River in the Linjiang City, Jilin Province, China (41.8087°N, 126.9211°E), and deposited at the Zoological Museum of Fudan University (Cuizhang Fu, [email protected]) under the voucher number FDZM-MeLLJ 20170922-01.
3. Methods
We extracted genomic DNA from muscle tissues following a high-salt protocol (Miller et al. Citation1988). The mitochondrial genome fragments were amplified using 13 primer pairs following a protocol previously described in our lab (Chai and Fu Citation2020; PCR gel image in Figure S1 of Appendix I). For the present study, we designed the forward primer of Primer9 (MeLND4F: 5′-TAGCCAGCCAAAAYCACAT-3′), as well as the reverse primers of Primer8 (MeLND4R: 5′-TAAAATCTGRTGGGCCGG-3′) and Primer10 (MeLND5R: 5′-AAACGRCTTGCCTGRGGAAG-3′). The remaining primers were adopted from Chai and Fu (Citation2020) (Table S1 in Appendix I). The PCR products were sequenced by the Sanger method using the ABI 3730 platform (Applied Biosystems, Foster City, USA; raw sequencing results in Appendix II).
Table 1. Characteristics of the mitochondrial genome of M. lachneri.
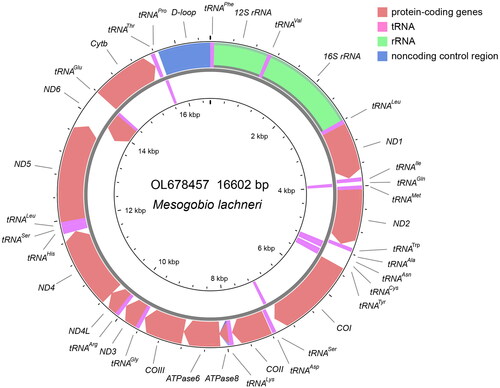
The contiguous and overlapping segments of M. lachneri were assembled with default parameters, using Sequencher 5.4 (Gene Codes, Ann Arbor, MI) on the basis of the reference genome of Gobio macrocephalus (MT632636; Yi and Fu Citation2020). MitoAnnotator was selected as the method for genome annotation (http://mitofish.aori.u-tokyo.ac.jp/annotation/input.html; Iwasaki et al. Citation2013). The CGView Server was used to generate the genome map (Grant and Stothard Citation2008).
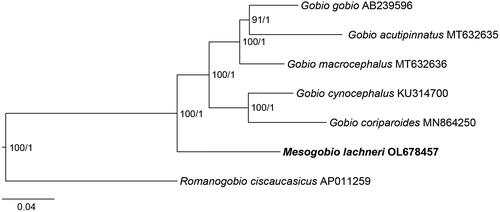
Tang et al. (Citation2011) have suggested that the genus Gobio is a close relative of the Mesogobio. Due to the fact that no other mitochondrial genome of fish belonging to the genus Mesogobio is available in GenBank, the mitochondrial genomes of five Gobio species (G. gobio, AB239596, Saitoh et al. Citation2006; G. acutipinnatus, MT632635, Yi and Fu Citation2020; G. macrocephalus, MT632636, Yi and Fu Citation2020; G. cynocephalus, KU314700, Li et al. Citation2018; and G. coriparoides, MN864250, Ge et al. Citation2020) and one Romanogobio species (R. ciscaucasicus, AP011259, Iwasaki et al. Citation2013) were downloaded from GenBank. The 13 protein-coding genes (PCGs) were used in the phylogenetic analyses, and R. ciscaucasicus was chosen as the outgroup taxon (Tang et al. Citation2011). For phylogenetic analyses, the data were partitioned by gene. The maximum-likelihood analysis was conducted using 1000 ultra-fast bootstrap replicates, and the best nucleotide substitution models were selected using the ‘-MF-mtree’ module- implemented in IQ-TREE v2.1.3 (Minh et al. Citation2020). Bayesian’s analysis was performed using two replicates of 50 million iterations, and the best models were automatically searched using the bModelTest module, implemented in BEAST v2.6.7 (Bouckaert and Drummond Citation2017).
4. Results
The mitochondrial genome of M. lachneri (OL678457) assembled from PCR products, presented as a circular structure with length of 16,602 bp, including a total of 37 genes (22 tRNA genes, 13 PCGs, and two rRNA genes) and one control region (). The overall base composition was composed of 30.1% A, 26.4% T, 17.1% G, and 26.5% C. For the 13 PCGs, two types of starting codons (ATG and GTG), and four kinds of stop codons (TAA, TAG, TA-, and T-) were observed (). Same patterns for codon use, are common in published mitochondrial genomes of the subfamily Gobioninae (Tong and Fu Citation2019; Zhang and Fu Citation2019; Fu and Fu Citation2020; Yi and Fu Citation2020). Reconstructed phylogenetic relationships between mitogenomes show that M. lachneri is the close relative of the genus Gobio with strong bootstrap support (100%, ).
5. Discussion and conclusions
The systematic position and status of the genus Mesogobio have remained unresolved to date, due to a lack of molecular data for the type species M. lachneri (Tang et al. Citation2011). In the present study, we determine the complete mitochondrial genome of M. lachneri for the first time. Our phylogenetic analysis reveals that M. lachneri has a close relationship with the genus Gobio, indicating that the genus Mesogobio may be a valid genus. The provided mitogenome of M. lachneri is expected to be useful for molecular species delimitation and for assessment of the molecular evolution of the family Gobionidae in the future.
Ethics statement
The sample of this study did not involve endangered or protected animals, and the experiment followed Laboratory animal—Guideline for ethical review of animal welfare of the National Standard of the People’s Republic of China (GB/T 35892-2018).
Author contributions
FCZ conceived this study; TW and NXM conducted the experiments, and analyzed the data; TW wrote the drafting of the paper; FCZ and NXM revised it critically, and that all authors agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (13.3 MB)Supplemental Material
Download MS Word (67.4 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The new genome in this study is openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/OL678457. Raw Sanger sequencing data are provided in supplementary material.
Additional information
Funding
References
- Bouckaert RR, Drummond AJ. 2017. bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol Biol. 17(1):42.
- Chai J, Fu CZ. 2020. Three mitochondrial genomes of freshwater fishes in the genus Squalidus (Cypriniformes: Gobionidae). Mitochondrial DNA B Resour. 5(3):3779–3781.
- Fu JW, Fu CZ. 2020. Three mitochondrial genomes of Pseudogobio fishes (Cypriniformes: Gobionidae). Mitochondrial DNA B Resour. 5(3):3046–3047.
- Ge YS, Cheng QQ, Yan YB, Duan XC, Wang T, Zhang YP, Lou ZY, Du YY. 2020. The complete mitochondrial genome sequence and phylogenetic position of Gobio coriparoides. Mitochondrial DNA B Resour. 5(1):808–809.
- Grant JR, Stothard P. 2008. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36(Web Server issue):W181–W184.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Yasunobu M, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. Mitofish and Mitoannotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540.
- Li YH, Cao K, Fu CZ. 2018. Ten fish mitogenomes of the tribe Gobionini (Cypriniformes: Cyprinidae: Gobioninae). Mitochondrial DNA B Resour. 3(2):803–804.
- Miller SA, Dykes DD, Polesky HF. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16(3):1215.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Saitoh K, Sado T, Mayden RL, Hanzawa N, Nakamura K, Nishida M, Miya M. 2006. Mitogenomic evolution and interrelationships of the Cypriniformes (Actinopterygii: Ostariophysi): the first evidence toward resolution of higher-level relationships of the world’s largest freshwater fish clade based on 59 whole mitogenome sequences. J Mol Evol. 63(6):826–841.
- Tan M, Armbruster JW. 2018. Phylogenetic classification of extant genera of fishes of the order Cypriniformes (Teleostei: Ostariophysi). Zootaxa. 4476(1):6–39.
- Tang KL, Agnew MK, Chen WJ, Hirt MV, Raley ME, Sado T, Schneider LM, Yang L, Bart HL, He SP, et al. 2011. Phylogeny of the gudgeons (Teleostei: Cyprinidae: Gobioninae). Mol Phylogenet Evol. 61(1):103–124.
- Tong J, Fu CZ. 2019. Four complete mitochondrial genomes of Saurogobio fishes (Cypriniformes: Gobionidae). Mitochondrial DNA B Resour. 4(2):2175–2176.
- Xie YH. 2007. Freshwater fishes in Northeast China. Shenyang: Liaoning Science and Technology Press.
- Yang JQ, He SP, Freyhof J, Witte K, Liu HZ. 2006. The phylogenetic relationships of the Gobioninae (Teleostei: Cyprinidae) inferred from mitochondrial cytochrome b gene sequences. Hydrobiologia. 553(1):255–266.
- Yi TY, Fu CZ. 2020. Two mitochondrial genomes of freshwater gudgeons in the genus Gobio (Cypriniformes: Gobionidae). Mitochondrial DNA B. 5(3):3072–3073.
- Yue PQ. 1998. Gobioninae. In: Chen YY, editor. Fauna Sinica, Osteichthyes: Cypriniformes (II). Beijing: Science Press.
- Zhang X, Fu CZ. 2019. Two complete mitochondrial genomes of Paraleucogobio fishes (Cypriniformes: Gobionidae). Mitochondrial DNA B Resour. 4(2):2173–2174.