Abstract
Leptopus chinensis (Bunge) Pojark is a medicinal herb endemic to China. In this study, its complete chloroplast (cp) genome was characterized with a discussion of its phylogenetic placement. The cp genome of L. chinensis was 154,600 bp long, with a double-stranded circular tetrad structure containing a small single-copy (SSC) region (17,717 bp), a large single-copy (LSC) region (83,159 bp), and a pair of inverted repeat (IR) regions (26,862 bp each). The overall GC content of the genome was 36.8% (LSC, 34.7%; SSC, 29.8%; IR, 42.3%). Phylogenetic analysis indicated that L. chinensis is a sister species to L. cordifolius.
Leptopus chinensis (Bunge) Pojark (1833), a medicinal herb widely distributed in many regions of China, is commonly used to treat diseases such as jaundice, gastritis, and edema (Liao et al. Citation2003). Through in vitro experiments with aqueous and ethanol extracts of L. chinensis on Eca-109 cells, L. chinensis extract was demonstrated to exert a strong inhibitory effect on tumor cells (Lu et al. Citation2000; Yue et al. Citation2000; Long et al. Citation2002), indicating its high medicinal potential in the treatment of cancer. The chloroplast (cp) genome plays a vital role in determining species identification, phylogenetic relationships, and germplasm diversity. However, to date, the cp genome of L. chinensis has not been reported, and current studies have mainly concentrated on its medicinal value and chemical activity. Consequently, the complete cp genome of L. chinensis was characterized in this study, providing important information for phylogenetic, genetic diversity, and evolutionary studies of this valuable herb.
Fresh leaves of L. chinensis were collected from Yeri Village, Benzilan Town, Deqin County, Diqing Tibetan Autonomous Prefecture, Yunnan Province, China (99°8′23.07″E, 28°24′3.94″N; altitude: 2485 m). Plant samples were collected in accordance with the wild plant protection regulations of the People’s Republic of China and were approved by the Forestry and Grassland Bureau of Yunnan Province. A voucher specimen (SWFU20210780MFY) was deposited in the herbarium of Southwest Forestry University, China (http://bbg.swfu.edu.cn/, Xiao Yu, email: [email protected]). Total DNA was extracted from dried plant leaf specimens using a modified cetyltrimethylammonium bromide method and purified (Doyle and Doyle Citation1987). The purified DNA sample was then used to prepare the DNA sequencing library, and high-throughput paired-end sequencing (2 × 150 bp) was performed on an Illumina HiSeq 4000 platform (Illumina, San Diego, CA) by Tianjin Novogene Biotechnology Co., Ltd. (Tianjin, China). Totally, 3 Gbp of raw sequencing data was retrieved, and was used to assemble the cp genome using GetOrganelle (Jin et al. Citation2020) with the following parameters: wordize = 102; base coverage = 171.44; and k = 75, 85, 95, 105, 115, and 127. Annotation of the resultant cp genome was done using the online tool CPGAVAS2 (Shi et al. Citation2019), and was further manually improved in Geneious Prime (Kearse et al. Citation2012). The annotated cp genome of L. chinensis was submitted to GenBank under the accession number ON050995.
The complete cp genome of L. chinensis is 154,600 bp in total length, with a double-stranded circular tetrad structure comprising a small single-copy (SSC) region with a length of 17,717 bp, a large single-copy (LSC) region with a length of 83,159 bp, and a pair of inverted repeat (IR) regions with a length of 26,862 bp. The overall GC content of the genome was 36.8% (LSC, 34.7%; SSC, 29.8%; IR, 42.3%), and those of A, T, C, and G were 31.24%, 31.99%, 18.68%, and 18.09%, respectively. A total of 128 coding genes, including one pseudogene, 82 protein-coding genes (PCGs), eight ribosomal RNA genes (rRNAs), and 37 transfer RNA genes (tRNAs), were annotated. Using the online software MISA-web (Beier et al. Citation2017) with default parameters, 82 SSRs were detected, of which 57, 14, 4, 7, and 0 were mono-, di-, tri-, tetra-, and pentanucleotide SSRs, respectively.
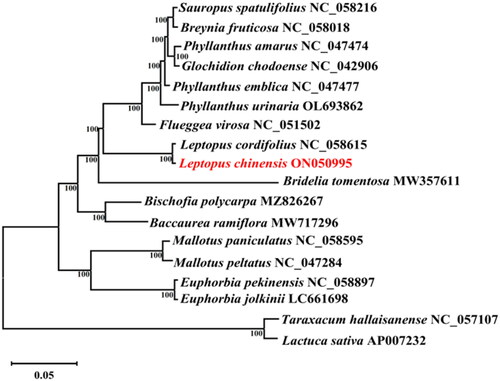
A phylogenetic tree was constructed based on the cp genome of L. chinensis and 15 species of Euphorbiaceae, with Taraxacum hallaisanense (NC_057107) and Lactuca sativa (AP007232) as the outgroup (). The MAFFT software (Katoh and Standley Citation2013) was used for multiple alignment among the cp genomes of these 18 species (scoring matrix = 200; PAM k = 2; gap open penalty = 1.53; offset value = 0.123). Subsequently, the resultant alignment was checked using MEGA v.11 software (Tamura Citation2021), and exported into the *.NET format. An maximum-likelihood (ML) phylogenetic tree was constructed using RAxML ver.8.0.0 (Stamatakis Citation2014) with the following parameters: bootstrap = 1000 and m = GTR + GAMMA. The phylogenetic tree was visualized using FigTree ver.1.4.3 software (http://tree.bio.ed.ac.uk/software/figtree/). The result revealed that all Euphorbiaceae species formed a monophyletic clade. In addition, L. chinensis and L. cordifolius were shown to be sister species. Our cp genome data of L. chinensis would contribute to future studies of taxonomy, genetics and genomics of related taxa.
Figure 1. ML phylogenetic tree based on complete chloroplast genome of L. chinensis and 17 other species. Numbers in the nodes are the bootstrap values from 1000 replicates. The following sequences were used: Sauropus spatulifolius NC_058216, Breynia fruticosa NC_058018 (Zhou et al. Citation2020), Phyllanthus amarus NC_047474, Glochidion chodoense NC_042906, Phyllanthus emblica NC_047477, Phyllanthus urinaria OL693862, Flueggea virosa NC_051502, Leptopus cordifolius NC_058615 (Rehman et al. Citation2021), Leptopus chinensis ON050995, Bridelia tomentosa MW357611, Bischofia polycarpa MZ826267, Baccaurea ramiflora MW717296 (Niu and Liu Citation2022), Mallotus paniculatus NC_058595, Mallotus peltatus NC_047284, Euphorbia pekinensis NC_058897, Euphorbia jolkinii LC661698 (Iwata et al. Citation2022), Taraxacum hallaisanense NC_057107 (Lee et al. Citation2021), and Lactuca sativa AP007232.
Author contributions
X. Y. conceived the study, collected the molecular materials; Z. N. Z. analyzed the experimental data and drafted the manuscript. Both authors provided comments and final approval.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that are newly obtained at this study are available in the NCBI under accession number of ON050995 (https://www.ncbi.nlm.nih.gov/nuccore/ON050995). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA820363, SRR18494790, and SAMN26994331, respectively.
Additional information
Funding
References
- Beier S, Thiel T, Münch T, Scholz U, Mascher M. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16):2583–2585.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf material. Phytochem Bull. 19:11–15.
- Iwata H, Ito T, Kokubugata G, Takayama K. 2022. The complete chloroplast genome of a coastal plant, Euphorbia jolkinii (Euphorbiaceae). Mitochondrial DNA Part B. 7(3):569–570.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lee W, Kim YS, Kim SC, Pak JH. 2021. The complete chloroplast genome sequence of a narrow alpine endemic, Taraxacum hallaisanense (Asteraceae), on Jeju Island, Korea. Mitochondrial DNA Part B. 6(3):1036–1038.
- Liao XC, Yu YZ, Chen XL, Lu JS, Qu LB, Zhao YF. 2003. Re-study of the chemical composition and crystal structure of Leptopus chinensis. Chin Trad Herb Drugs. 9:24–25.
- Long Y, Lu JS, Chen XL. 2002. The analysis of volatile oils of Leptopus chinensis (Bunge) Pojark. J Chin Med Mater. 25(3):181–182.
- Lu J, Long Y, Chen X. 2000. The elementary analysis on herbal medicine Leptopus Chinensis (Bunge) Pojark. J Hennan Med Univ. 35(1):53–54.
- Niu YF, Liu J. 2022. Complete chloroplast genome of Baccaurea ramiflora and its phylogenetic analysis. Mitochondrial DNA Part B. 7(1):206–207.
- Rehman U, Sultana N, Jamal A, Muzaffar M, Poczai P. 2021. Comparative chloroplast genomics in Phyllanthaceae species. Diversity. 13(9):403.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027.
- Yue L, Xiaolan C, Jiansha L, Weimin Z, Qingzhi Z. 2000. Determination of the content of amino acids in Leptopus chinensis (Bunge) Pojark. J Henan Med Univ. 35(4):325–326.
- Zhou Z, Cao R, Hu D, Liu J. 2020. Characterization of the complete plastid genome sequence of Breynia fruticosa (L.) Müll. Arg. (Phyllanthaceae), a traditional Chinese medicine plant. Mitochondrial DNA Part B. 5(3):3510–3511.
