Abstract
In this study, we analyzed the complete mitochondrial genome of Eudynamys scolopaceus using Illumina and Pacbio next-generation sequencing. The total length of the mitogenome was 18,170 bp and contained 13 protein-coding genes (PCGs), two ribosomal RNAs, 22 transfer RNAs, a non-coding control region, and two repeat regions. The order of the genes and genome structure were the same as those in a previous publication in other Cuculidae species. A phylogenetic tree constructed with 13 PCGs indicated that E. scolopaceus is closely related to Urodynamis taitensis and genus Cuculus.
The Asian Koel (Eudynamys scolopaceus Linnaeus, 1758) is a large cuckoo species in the family Cuculidae. The main phenotypic characteristic of male is a glossy bluish-black body, a long tail, and a crimson iris. The female has a dark brown body with many pale spots and a long tail. E. scolopaceus is distributed in Central and Southern China, the Indochina Peninsula, and Southeast Asia (Won and Kim Citation2012). In South Korea, E. scolopaceus are occasionally observed as vagrant birds. According to the International Union for Conservation of Nature (IUCN) Red List, E. scolopaceus is listed as a least concern (LC) species (BirdLife International Citation2016). Mitochondrial genome is valuable for analyzing the phylogenetic relationships and evolutionary status of species. The complete mitochondrial genome of E. scolopaceus had not been published from South Korea. In this study, we describe the complete mitochondrial genome of E. scolopaceus from South Korea and explore the phylogenetic relationships among species of Cuculidae.
E. scolopaceus (NIBRGR0000637157) was collected from Baengnyeong Island, Ongjin-gun, Incheon, South Korea (37° 57′ 36.6″N, 124° 39′ 53.3″E) on 21 May 2020 as a carcass. Sample collection was carried out with the approval of a wild animal collection permit (no. 2020-1 Ongjin-gun) and the specimen was identified by Dong-won Kim (National Migratory Birds Center, Incheon, Republic of Korea). Muscle tissue samples for the experiment were taken from the chest and stored in the deep-freezer of the National Migratory Birds Center, National Institute of Biological Resources (NIBR), Incheon until DNA extraction (in charge of the samples: Dong-won Kim, [email protected]).
Total genomic DNA was extracted using the QIAamp DNA Micro Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Next-generation sequencing was carried out on the Illumina HiseqX and Pacbio RSII platform at Geno Tech Corp. (Daejeon, South Korea). De novo assembly of the mitogenome was performed using CLC Genomics Workbench v6.5.1 (CLCbio, Aarhus, Denmark) and SMRT Analysis v2.3.0 HGAP.2 (Chin et al. Citation2013). Gene structures of the mitochondrial genome were annotated with MITOS web program (Bernt et al. Citation2013). The complete mitochondrial genome sequence of E. scolopaceus was submitted to GenBank (accession no. OL639163).
The mitogenome sequence of E. scolopaceus was 18,170 bp in length and contained 13 protein-coding genes (PCGs), two ribosomal RNA genes (rRNAs), 22 transfer RNA genes (tRNAs), and a non-coding control region (CR) with two repeat regions, similar to other Cuculidae species. The G-C content (41.9%) was significantly lower than the A-T content (58.1%) with the following base-pair composition: 34.1% (A), 29.6% (C), 12.3% (G), and 24.0% (T). Nine of the genes in E. scolopaceus were located on the light strand (L-strand), including one PCG (ND6) and eight tRNA genes (tRNA-Gln, Ala, Asn, Cys, Tyr, Ser, Pro, and Glu). The other 29 genes were located on the heavy strand (H-strand). Nine PCGs started with ATG; the exceptions were COX1 (GTG), ND2, ND3, and ND5 (ATA). Ten PCGs terminated with the complete vertebrate mitochondrial stop codons TAA (COX2, ATP8, ATP6, ND3, ND4L, CYTB, and ND5), AGG (COX1), AGA (ND1), and TAG (ND6). Only three genes had incomplete stop codons, ending as ATT (ND2), CTT (COX3), and TAT (ND4).
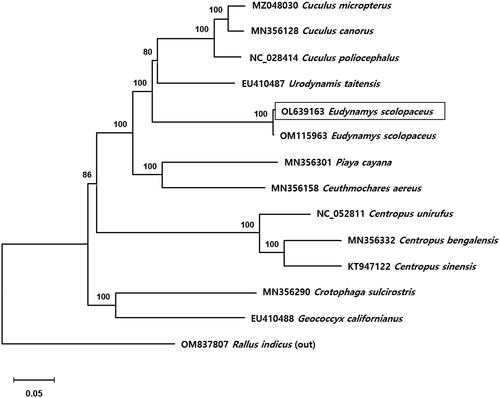
The phylogenetic maximum-likelihood (ML) tree based on the 13 concatenated PCGs was constructed using MEGA X (Kumar et al. Citation2018) with 1000 bootstrap replicates. The most appropriate models for the ML trees were selected with MEGA X (Kumar et al. Citation2018). The best-fit model for the ML tree was the general time reversible (GTR) model with Gamma distributed (+G) and proportion of invariant sites (+I). The mitochondrial genome reference for the phylogenetic tree corresponded to 12 individuals (12 species) of Cuculidae from GenBank. The Brown-cheeked Rail (Rallus indicus) was used as an out-group (OM837807). The phylogenetic result indicated that E. scolopaceus belongs to the family Cuculidae, and closely related to the Pacific Long-tailed Cuckoo (Urodynamis taitensis) and genus Cuculus (). U. taitensis was included in the genus Eudynamys in the past (Mayr Citation1944; Mason Citation1997), but it is now separated into a new genus, Urodynamis (Payne Citation2005). Also, phylogenetic tree showed that other species in the order Cuculiformes clustered within family groups. This phylogenetic tree topology was identical with recent study from South China (Wang et al. Citation2022). The mitochondrial genome of E. scolopaceus (OM115963) published by Wang formed a single clade with E. scolopaceus (OL639163 in this study) in the phylogenetic tree. In conclusion, our results provide a mitogenome database for further evolutionary and molecular research on Cuculidae.
Author contributions
All authors contributed to the concept and design of the study. Sample collection and preparation of materials were done by Eujin Cheong, Jin Young Park, Wee-Haeng Hur, Yu-Seong Choi, Hyung-Kyu Nam, and Dong-won Kim. Yun Sun Lee performed the data analysis, interpretation of the data, and wrote the first draft of the manuscript. All authors read and revised the manuscript and approved the final version. All authors agree to be accountable for all aspects of the paper.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OL639163. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA815876, SRR18319842, and SAMN26646336, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- BirdLife International. 2016. Eudynamys scolopaceus: the IUCN Red List of Threatened Species 2016; [accessed 2022 October 18]. https://dx.doi.org/10.2305/IUCN.UK.2016-3.RLTS.T22684049A93012559.en.
- Chin C, Alexander DH, Marks P, Klammer AA, Drake JP, Heiner CR, Clum A, Copeland A, Huddleston J, Eichler EE, et al. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 10(6):563–569.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Mason IJ. 1997. Cuculidae, Centropidae. In: Houston WWK, Wells A, editors. Zoological catalogue of Australia: Aves (Columbidae to Coraciidae). Melbourne: CSIRO; p. 255–263.
- Mayr E. 1944. Birds collected during the Whitney South Sea Expedition 54. Notes on some genera from the Southwest Pacific. Am Mus Novit. 1269:1–8.
- Payne RB. 2005. Bird families of the world: Cuckoos. England: Oxford University Press.
- Wang G, Tang C, Li J, Huang Q, Nong J, Wei L, Zhou Q. 2022. Complete mitogenome of the Common Koel Eudynamys scolopaceus Linnaeus 1758 (Aves: Cuculidae). Mitochondrial DNA Part B. 7(5):831–833.
- Won BO, Kim HJ. 2012. The birds of Korea. Seoul: Academy Book; p. 524–525.