Abstract
Carallia diplopetala (Rhizophoraceae) is an important economic tree species narrowly distributed endemic to East Asia. In this study, We generate the complete chloroplast genome of C. diplopetala using next-generation sequencing technology, which is 162,052 bp in size and consists of a large single copy (LSC) of 89,556 bp and a small single copy (SSC) of 18,814 bp, separated a pair of inverted repeats (IRb and IRa) of 26,841 bp. The overall GC content is 36.4%. A total of 130 genes are annotated, including 83 protein-coding genes, 37 tRNAs, eight rRNAs and two pseudogenes (ψycf1 and ψrps19). The phylogenetic analysis indicated that C. diplopetala and C. brachiate formed a monophyletic clade with strong support and then sister to Pellacalyx yunnanensis. The plastome of C. diplopetala will provide informative genomic resources for further conservation applications.
Carallia diplopetala Hand.-Mazz. 1931 belongs to the mangrove family (Rhizophoraceae) and is an endemic species in East Asia with narrow distribution in China and Vietnam (Qin and Boufford Citation2007). This species provides a higher quality timber for furniture and industrial purposes (Liang et al. Citation2010). Its roots and leaves have been consumed as herbs in traditional Chinese medicine, and they are effective in treating rheumatism, bleeding and fever (Xiao and Wang Citation1992). With long-term over-excavation of the wild resources, the habitat has been severely fragmented and the populations have decreased significantly. Therefore, it was listed as an endangered species in China Species Red List (Fu Citation1992), requiring urgent conservation and restoration. However, there’s limited genomic information in regard to C. diplopetala. Here, we sequenced the complete chloroplast (cp) genome sequence of C. diplopetala, which will provide vital genetic information for sustainable management and utilization of this species.
The fresh leaves of C. diplopetala were collected in Guangxi Fangcheng Golden Camellias National Nature Reserve, Guangxi Province, China (21°45'37"N, 108°5′ 27"E). These samples collection was approved by the local management department. Total genomic DNA was extracted from silica-dried leaves by modified CTAB (hexadecyltrimethylammonium bromide) method (Doyle and Doyle Citation1987). A specimen was deposited at Guangxi Forestry Research Institute (contact: Ronglin Wang, e-mail: [email protected], http://www.gxlky.com.cn/) under the voucher number hrl20210906001. The isolated genomic DNA was subsequently fragmented in length of 350 bp and sequenced on an Illumina Hiseq X-ten platform (San Diego, USA) at Novogene (Beijing, China). A total of 2 Gb raw Paired-end reads were yielded and de novo assembled into chloroplast genome using NOVOPlasty 4.3.1 (Dierckxsens et al. Citation2017). The genome annotation was performed by GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html; Tillich et al. Citation2017) and adjusted by manual in Geneious 11.1.5 (Kearse et al. Citation2012). The complete sequence data was submitted to National Center for Biotechnology Information (NCBI) under the accession number OM001094.
The chloroplast genome of C. diplopetala exhibited a typical double-stands circular structure with 162,052 bp in size, consisting of two duplicate inverted repeats (IRb and IRa) of 26,841 bp, isolated by the large and small single copy (LSC and SSC) regions of 89,556 bp and 18,814 bp, respectively. The overall GC content of plastome was 35.8%, with the GC contents of LSC, SSC and IRs at 33.2%, 29.6%, and 42.4%, respectively. A total of 130 genes were annotated, including 83 protein-coding genes, 37 transfer RNA genes (tRNA), eight ribosomal RNA genes (rRNA) and two pseudogenes (ψycf1 and ψrps19). Among them, six protein-coding genes (rpl2, rpl23, ycf2, ndhB, rps7 and rps12), seven tRNA (trnI-GAU, trnA-UGC, trnL-CAA, trnR-ACG, trnV-GAC, trnN-GUU and trnM-CAU), and four rRNA (4.5S, 5S, 16S and 23S rRNA) were duplicated in the IR regions.
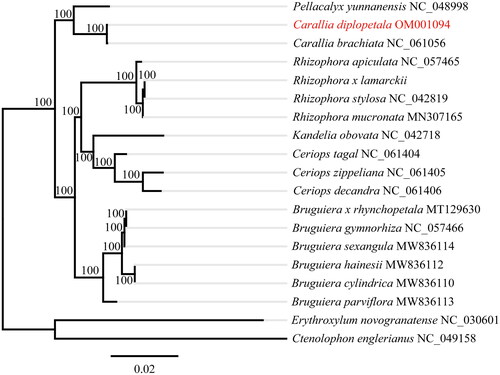
To reveal the phylogenetic relationship of C. diplopetala within Rhizophoraceae, we downloaded additional 17 species of Rhizophoraceae from Genbank, two species from Ctenolophonaceae (Ctenolophon englerianus, NC_049158) and Erythroxylaceae (Erythroxylum novogranatens, NC_030601) were chosen as outgroups according to the result of Xi et al. (Citation2012) and Ruang-areerate et al. (Citation2022). All sequences were aligned by MAFFT 7.409 (Katoh and Standley Citation2013). The phylogenetic analysis was conducted under the GTR + GAMMA model in RAxML with 1000 bootstrap replicates (Stamatakis Citation2014). Maximum likelihood (ML) phylogenetic tree indicated that C. diplopetala and C. brachiate formed a monophyletic clade with strong support and then sister to Pellacalyx yunnanensis (). The topology of Rhizophoraceae is largely congruent with the result indicating from transcriptomes data (Guo et al. Citation2017).
Figure 1. Phylogenetic tree of Carallia diplopetala using maximum likelihood method. Numbers on each node indicated the bootstrap support values after 1000 replicates. The following sequences were used: Ceriops tagal NC_061404, Ceriops zippeliana NC_061405, Ceriops decandra NC_061406 (Ruang-areerate et al. Citation2022); Pellacalyx yunnanensis NC_048998 (Zhang et al, Citation2019); Kandelia obovata NC_042718 (Chen et al. Citation2019); Rhizophora stylosa NC_042819 (Li et al. Citation2019); Rhizophora x lamarckii NC_046517, Rhizophora stylosa NC_042819, Rhizophora mucronate MN307165, Rhizophora apiculate NC_057465, Bruguiera cylindrica MW836110, Bruguiera hainesii MW836112, Bruguiera parviflora MW836113, Bruguiera sexangular MW836114, Bruguiera gymnorhiza NC_057466, Carallia brachiate NC_061056, Carallia diplopetala OM001094 (this study), Erythroxylum novogranatense NC_030601 and Ctenolophon englerianus NC_049158 (Wang et al. Citation2019).
Author contributions statement
Renjie Wang, Nanyan Liao, Yi Qin, Yong Wang and Ronglin Huang participated in the study design, data collection, analysis of data and preparation of the manuscript. Xiongsheng Liu, Ronglin Huang and Yufei Xiao revised this paper critically for important intellectual content. All authors agree to be accountable for all aspects of the work.
Disclosure statement
The authors declare no conflict of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession number OM001094. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA792453, SRR17332993 and SAMN24425643, respectively.
Additional information
Funding
References
- Chen D, Xiang S, Liu Z, Zou S. 2019. The complete chloroplast genome sequence of Kandelia obovata (Rhizophoraceae). Mitochondrial DNA Part B. 4(2):3494–3495.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fu L. 1992. China plant red data book: rare and endangered plants. Vol. 1. Beijing: Chinese Science Press.
- Guo W, Wu H, Zhang Z, Yang C, Hu L, Shi X, Jian S, Shi S, Huang Y. 2017. Comparative analysis of transcriptomes in Rhizophoraceae provides insights into the origin and adaptive evolution of mangrove plants in intertidal environments. Front Plant Sci. 8:795.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li C, Guo P, Huang H, Pei N, Shi M, Yan H. 2019. The complete chloroplast genome of Rhizophora stylosa and its phylogenetic implications. Mitochondrial DNA Part B. 4(1):374–375.
- Liang H, Wei J, Wang M, Li H, Jiang S. 2010. Photosynthetic characteristics of young and mature Carallia diplopetala. J Fujian Fore Sci Tech. 37:51–54.
- Qin HN, Boufford DE. 2007. Carallia diplopetala. In Wu ZY, Raven PH, Hong DY, editors. Flora of China Volume 13 (Rhizophoraceae). Beijing & St. Louis: Science Press & Missouri Botanical Garden Press; p. 298–299.
- Ruang-areerate P, Yoocha T, Kongkachana W, Phetchawang P, Maknual C, Meepol W, Jiumjamrassil D, Pootakham W, Tangphatsornruang S. 2022. Comparative analysis and phylogenetic relationships of Ceriops species (Rhizophoraceae) and Avicennia lanata (Acanthaceae): insight into the chloroplast genome evolution between middle and seaward zones of mangrove forests. Biology. 11(3):383.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang Z, Jin D, Wang G, Yi T. 2019. The complete plastome of Ctenolophon englerianus Mildbr. Mitochondrial DNA Part B. 4(2):3379–3380.
- Xi Z, Ruhfel BR, Schaefer H, Amorim AM, Sugumaran M, Wurdack KJ, Endress PK, Matthews ML, Stevens PF, Mathews S, et al. 2012. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc Natl Acad Sci USA. 109(43):17519–17524.
- Xiao W, Wang H. 1992. A tree with multi-uses – Carallia lanceaefolia Roxb. Chin Wild Plant Res. 4:23–24.
- Zhang J, Li Y, Yuan X, Wang Y. 2019. The complete chloroplast genome sequence of Pellacalyx yunnanensis: an endangered species in China. Mitochondrial DNA Part B. 4(2):3948–3949.
