Abstract
Hibiscus trionum L. is an annual herbaceous plant belonging to the Malvaceae family. It is native to Central Africa, however, is now naturalized in Europe and Asia including Korea. Here, we report the complete chloroplast genome assembly of H. trionum. The complete chloroplast genome comprises 160,530 bp and is divided into four typical regions: a large single-copy region of 89,272 bp, a pair of inverse repeats of 26,152 bp each, and a small single-copy region of 18,954 bp. A total of 131 genes were identified in this chloroplast, of which 86 were protein-coding, 37 were tRNA, and 8 were rRNA genes. The results of this study will serve as a key reference for further research on Hibiscus speciation.
Hibiscus trionum Linnaeus 1753 is an annual herbaceous plant belonging to the family Malvaceae. H. trionum, originally native to Central Africa, is now considered naturalized in Europe and Asia (Craven et al. Citation2011). H. trionum is an important summer weed used in grain- and cotton-growing systems in the subtropical region of northeastern Australia (Walker et al. Citation2010). In China, it is grown as a common weed, and its fruits and seeds are used as antipyretics (Zhou et al. Citation2021). In this study, the chloroplast genome of H. trionum was assembled to understand its evolution in the gene migration process, with from endemic native to global distribution.
Fresh leaves of H. trionum were collected from an experimental site in the National Institute of Forest Science (Suwon, Korea; 37.15° N, 126.57° E). The specimen was deposited at the Korea National Arboretum (http://nature.go.kr, Hyuk-Jin Kim, [email protected]) under voucher number KNKA200005243003. Subsequently, total genomic DNA was extracted using a GeneAll Genomic DNA Purification Kit (GeneAll Biotechnology, Seoul, Korea). Next-generation sequencing library preparation was conducted by Macrogen Inc. (Seoul, Korea), using a TruSeq Nano DNA Kit (Illumina, San Diego, CA, USA). Genome sequencing was performed using a NovaSeq 6000 platform (Illumina). The chloroplast genome sequence was assembled using the organelle assembler software NOVOPlasty 4.3.1 (Dierckxsens et al. Citation2017), with H. sinosyriacus (MZ367751) as a reference sequence. In total of 143,520,816 reads, 13,719,462 reads mapped to the reference genome. Finally, a total of 2,785,690 reads were assembled, and the average organelle coverage was 12,905-fold.
The chloroplast genome sequence of H. trionum was deposited in NCBI GenBank (accession number OL628829). The chloroplast genome of H. trionum comprised 160,530 bp, divided into four regions: a large single-copy region (89,272 bp), two inverted repeats (26,152 bp, each), and a small single-copy region (18,954 bp). Furthermore, a total of 131 genes were identified, comprising 86 protein-coding sequences, and 37 tRNAs and 8 rRNAs genes.
For phylogenetic analyses, 79 protein sequences from each chloroplast genome of 18 species were used; these species belonged to four genera of the Malvaceae family, i.e. Hibiscus, Gossypium, Abelmoschus, and Talipariti. Analyses were performed using Clustal Omega 1.2.4 (Sievers et al. Citation2011). A phylogenetic tree of the aligned sequences was constructed using the maximum likelihood method and the JTT matrix-based model (Jones et al. Citation1992). A bootstrap consensus tree was inferred from 1,000 replicates using MEGA 11 software (Felsenstein Citation1985; Kumar et al. Citation2018). Phylogenetic analyses based on comparison of protein sequences showed that the genus Gossypium first diverged from other genera and occurred as an outgroup. H. trionum showed a paraphyletic relationship with several closely related species ().
Figure 1. Phylogenetic tree showing taxonomic relationships of Hibiscus trionum (in bold) with other species of the Malvaceae family (NCBI accession numbers are shown). Phylogenetic analyses were conducted with protein sequences using MEGA X. The bootstrapping tree was inferred using 1000 replicates.
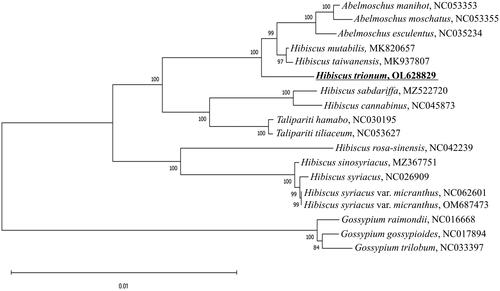
These phylogenetic insights based on the complete chloroplast genome of H. trionum will help interpret the evolution of this species and the relationships between its closely related species.
Author contributions statement
Soon-Ho Kwon contributed to the conceptualization, design, data analysis and interpretation, and drafting of the manuscript. You Lim Jang contributed to sample collection and drafting of the manuscript. Hae-Yun Kwon contributed to the design of the study, and drafting, and revising the final version of the manuscript. All authors approved the final version and agreed to be accountable for all aspects of the work.
Ethics statement
Based on Article 15: Exceptions to the Breeder’s Right to the International Convention for the Protection of New Varieties of Plans (UPOV Publication. No. 221(E) Article 15: (1)-ii) of the International Union for the Protection of New Varieties of Plants (UPOV) in 1991, this study can be conducted without ethical approval or permission.
Disclosure statement
The authors have no relevant financial or non-financial competing interests to declare.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/ under accession no. OL628829. The associated BioProject, BioSample, and SRA numbers are PRJNA798560, SAMN25084885, and SRR17686673, respectively.
Additional information
Funding
References
- Craven LA, de Lange PJ, Lally TR, Murray BG, Johnson SB. 2011. A taxonomic re-evaluation of Hibiscus trionum (Malvaceae) in Australasia. NZ J Bot. 49(1):27–40.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39(4):783–791.
- Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 8(3):275–282.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7(539):539.
- Walker S, Wu H, Bell K. 2010. Emergence and seed persistence of Echinochloa colona, Urochloa panicoides and Hibiscus trionum in the sub-tropical environment of north-eastern Australia. Plant Prot. Q. 25:127–132.
- Zhou SX, Zhu ZX, Wei CX, Shi K, Han CX, Zhang C, Shao H. 2021. Chemical profile and phytotoxic action of Hibiscus trionum essential oil. Chem Biodivers. 18(2):e2000897.
