Abstract
This work determined and analyzed the complete chloroplast genome sequence of Ceratopteris thalictroides (Linnaeus) Brongniart 1822 (Pteridaceae). The results indicate that the total chloroplast genome size of C. thalictroides is 149,399 bp in length, and the genome contains a large single-copy (LSC) region of 83,580 bp, a small single-copy (SSC) region of 21,241 bp, and a pair of inverted repeat (IR) regions of 22,289 bp. The GC content of C. thalictroides is 36.7%. The genome encodes a total of 131 unique genes, including 82 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The phylogenetic analysis results strongly suggest that C. thalictroides is closely related to C. cornuta.
The water fern Ceratopteris thalictroides (Linnaeus) Brongniart 1822 grows in marshlands and paddy fields and sometimes floats on the water. It was previously classified as a member of Parkeriaceae (Ching Citation1978) but is now considered a member of Pteridaceae, which is located at the base of the order Polypodiales on the phylogenetic tree (Smith et al. Citation2006; PPG I Citation2016; Shen et al. Citation2018). It is widely distributed worldwide throughout the tropics. Because of the destruction of its habitat and extensive plundering for its edible, ornamental, and medicinal properties, C. thalictroides is treated as a second-class protected wild plant in China (National Forestry Administration of the People’s Republic of China & Ministry of Agriculture of the People’s Republic of China Citation2021). C. thalictroides is an excellent model plant because of its short lifespan and diverse mechanisms of reproduction (Li and Wang Citation1997). To date, no studies on the complete chloroplast genome of C. thalictroides have been published. Therefore, we analyzed the complete chloroplast genome of C. thalictroides in this study for subsequent molecular phylogenetic analysis.
The C. thalictroides sample used in this study was collected from Guantouzui, Hanshou County, Hunan Province, China (111, 58′ 32.0641″ E; 29, 1′ 39.1464″ N; altitude: 9 m); the specimen was deposited at the Chenshan Herbarium (CSH, Shanghai, China) and assigned voucher number Fern09721, Zhou XL, 2018 (Zhang Rui, [email protected]).
The genomic DNA was extracted from the silica gel dried leaf of C. thalictroides and sequenced on the Illumina HiSeq platform (Shanghai Majorbio Bio-pharm Technology Co., Ltd., Shanghai, China). The plastid genome was assembled using GetOrganelle (Jin et al. Citation2018) with the chloroplast genome of C. cornuta (accession number: MH173068) as the reference sequence. The assembled chloroplast genome was annotated by Geneious Prime (Biomatters Ltd., Auckland, New Zealand) (Kearse et al. Citation2012). Finally, the complete chloroplast genome of C. thalictroides was obtained; the genome sequence data are openly available in the GenBank database of the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov/ under accession number OK524221.
The results indicate that the chloroplast genome size of C. thalictroides (OK524221) is 149,399 bp in length, and the genome contains a large single-copy (LSC) region of 83,580 bp, a small single-copy (SSC) region of 21,241 bp, and a pair of inverted repeat (IR) regions of 22,289 bp. The GC content of C. thalictroides is 36.7%. The genome encodes a total of 131 unique genes, including 82 protein-coding genes, 38 tRNA genes, and 8 rRNA genes.
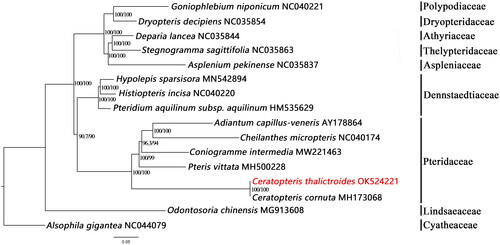
For the molecular phylogenetic analysis, the complete chloroplast genomes of 15 ferns were downloaded from GenBank to obtain a phylogenetic tree containing C. thalictroides. The sequences were aligned by MAFFT (Katoh and Standley Citation2013). The phylogenetic tree was constructed using the IQ-TREE (maximum likelihood) method in PhyloSuite, and branch supports were constructed with ultrafast bootstrap approximation (Guindon et al. Citation2010; Minh et al. Citation2013; Nguyen et al. Citation2015; Zhang et al. Citation2020). The results revealed that C. thalictroides is a sister of C. cornuta MH173068 (). This is consistent with previous studies on the Pteridaceae phylogenetic tree.
Ethical approval statement
Research on plant chloroplast genome sequencing does not require ethical approval.
Author contribution statement
Liu Xing-Feng (corresponding author and first author): resources, conceptualization, investigation, and writing—original draft. Zhou Xi-Le (first author): investigation, funding acquisition, resources, supervision, writing—review & editing. Gu Yu-Feng: data curation, methodology, and visualization. Liu Si-Fan: resources, formal analysis, and validation. Yu Jun-Hao: data curation. Zhang Rui: funding acquisition and methodology. Shu Jiang-Ping: software. All authors have read and agreed to the published version of the manuscript.
Plant material collection statement
The species is widely distributed in China; the plant materials were not obtained from nature reserves, and the chloroplast genome sequencing research did not affect the population, so collection licenses were not needed.
Disclosure statement
The authors report no conflicts of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in the GenBank database of NCBI at https://www.ncbi.nlm.nih.gov (https://www.ncbi.nlm.nih.gov/nuccore/OK524221.1/) under accession no. OK524221. The associated BioProject, Sequence Read Archive, and BioSample numbers are PRJNA733526, SRR14683085, and SAMN19414834, respectively.
Additional information
Funding
References
- Ching RC. 1978. The Chinese fern families and genera: systematic arrangement and historical origin. J Systemat Evol. 16(4):16–37.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 256479. doi:10.1101/256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li JY, Wang TX. 1997. Ceratopteris thalictroides is a good material for studying plant genetics. Bull Biol. 32(10):40.
- Minh BQ, Nguyen MA, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- PPG I. 2016. A community-derived classification for extant lycophytes and ferns. J System Evol. 54(6):563–603.
- Shen H, Jin D, Shu JP, Zhou XL, Lei M, Wei R, Shang H, Wei HJ, Zhang R, Liu L, et al. 2018. Large scale phylogenomic analysis resolves a backbone phylogeny in ferns. Gigascience. 7(2):1–11.
- Smith AR, Pryer KM, Schuettpelz E, Korall P, Schneider H, Wolf PG. 2006. A classification for extant ferns. Taxon. 55(3):705–731.
- The Natioal Forestry Administration of the People’s Republic of China & the Ministry of Agriculture of the People’s Republic of China. 2021. List of national key protected wild plants.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.