Abstract
Veronica arvensis, which is an annual flowering plant in the plantain family Plantaginaceae, has commonly used as a Chinese herbal medicine to treat malaria in China. Here, the complete plastome of V. arvensis was successfully assembled based on genome skimming sequencing. The plastome of V. arvensis was 149,386 bp in length, comprising a pair of inverted repeats (IR; 24,946 bp) separated by a large single-copy (LSC) region (82,004 bp) and a small single-copy (SSC) region (17,490 bp). The plastid genome encoded 113 unique genes, consisting of 79 protein-coding genes, 30 tRNA genes, and four rRNA genes, with 19 duplicated genes in the IR regions. Six plastid hotspot regions (trnH-psbA, trnK-rps16, atpI-rps2, ndhF-rpl32, ccsA-ndhD and rps15-ycf1) were identified within Veronica. Phylogenetic analysis showed that the representative species from Veronica was monophyletic. V. persica and V. polita formed a maximum clade, followed by sister to V. arvensis.
For the past three decades, molecular data have been increasingly used to characterize plant diversity, which has had a profound impact on our understanding of plant diversification patterns and phylogenetic relationships (Straub et al. Citation2012). Plastomes, usually mapped as circular genomes, are increasingly used for reconstructing the angiosperm phylogenetic backbone due to its relatively small size, conserved gene content and simple structure (Liu et al. Citation2017). Recent advances in sequencing technology have made it easier to obtain the complete plastomes for any plant species, regardless of model or not model organisms (Malé et al. Citation2015). Veronica L. is the largest genus in the flowering plant family Plantaginaceae, with about 500 species distributed in the world (Mazur et al. Citation2021). However, there were only six plastomes sequenced and submitted to GenBank (retrieved July 2022 from https://www.ncbi.nlm.nih.gov), which hinder to resolve the phylogeny of Veronica. V. arvensis Linaeus 1753 (), which is native to European, is an annual flowering plant of Veronica and usually grown in gardens, pastures, and cultivated land (Baskin and Baskin Citation1983). According to traditional Chinese medicine, whole plant of V. arvensis had been used to treat malaria in Jiangsu and Zhejiang provinces (Zhao Citation1963). In this study, the plastid genome of V. arvensis was firstly sequenced and assembled using genome skimming data, and comparative analyses of plastomes were performed for several representative species to screen the highly divergent regions (plastid hotspot) in the plastome of Veronica species.
Figure 1. The morphological characteristics of V. arvensis. A, B, C showing the photo of whole plant, flowers, and fruits, respectively (photos taken by Qi Li at Henan University).

Fresh and healthy leaves of V. arvensis were collected in Jinming campus of Henan University, Kaifeng (China; 114°18′14.23″E, 34°49′08.72″N). Field work for collecting plant sample did not require a specific permission, and no protected or endangered species were involved. A voucher specimen (LQ20220401; Qi Li, [email protected]) was deposited in the Herbarium of Henan University (HHN). Total genomic DNA was extracted using Plant DNAzol Reagent (LifeFeng, Shanghai) according to the manufacturer’s protocol with ∼6 mg of the fresh leaf tissue. Illumina paired-end libraries (150 bp read length) were prepared from sheared genomic DNA (fragment size ≤800 bp) and sequenced on an Illumina HiSeq X10 by Beijing Genomics Institute (BGI, Wuhan, China). The raw reads with the adapters trimmed were filtered by quality with Phred scores of 30 or less implemented in the CLC-quality trim tool, and the filtered reads were assembled using GetOrganelle pipeline (Jin et al. Citation2020). Combined commands were employed in the pipeline to recruit plastid-like reads using Bowite2 (Langmead and Salzberg Citation2012) and assembled them using SPAdes (Bankevich et al. Citation2012). The software Geneious R11 (Biomatters, Auckland, New Zealand) was used to annotate the plastome with V. eriogyne H. Winkl. 1922 (GenBank accession number: MZ323109) as a reference. The plastome map of V. arvensis was drawn using the OGDRAW program (Lohse et al. Citation2013). The genome sequence was registered into GenBank with the accession number ON461915. Multiple alignments of the seven plastomes of Veronica species (the species name and GenBank accession numbers were seen in ) were carried out using MAFFT v7.017 (Katoh and Standley Citation2013) to screen for plastid hotspot regions in Veronica, and nucleotide diversity (Pi) was determined using DnaSP v.5.0 (Librado and Rozas Citation2009). Maximum likelihood (ML) was performed to estimate the phylogeny for 27 plastome sequences of Plantaginaceae species, with two Osmanthus Lour. species as outgroups. ML analysis was implemented at the CIPRES Science Gateway v3.3 (Miller et al. Citation2010) using RAxML v8.1.11 with the optimal substitution model (GTR + I+G) determined by the software jModel Test v2.1.4 (Posada Citation2008), 1000 bootstrap iterations were conducted with other parameters using the default settings.
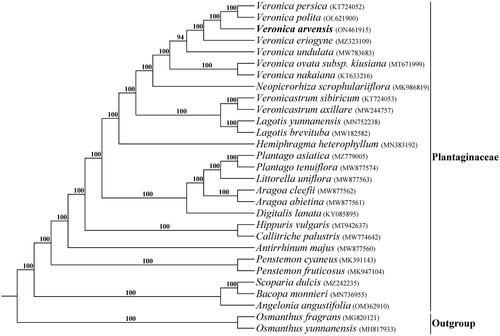
The total of 30,763,654 paired-end reads were generated from genome skimming sequencing for V. arvensis, and 7,053,345 reads were trimmed due to low quality. The plastome of V. arvensis was 149,386 bp in length and shared the common quadripartite structure of most angiosperms (). The genome comprised two copies of IR (24,946 bp each) separated by a LSC region (82,004 bp) and a SSC region (17,490 bp). There were 113 unique genes including 79 protein-coding genes, 30 tRNA genes and four rRNA genes annotated in the plastome of V. arvensis, with 19 duplicated genes in the IR regions. In addition, we generated 131 loci (70 genic and 61 inter-genic spacer regions) within Veronica with more than 200 bp in length and the nucleotide variability (Pi) values were ranged from 0.0009 to 0.1367 (). Six of these variable loci (Pi > 0.09) including trnH-psbA, trnK-rps16, atpI-rps2, ndhF-rpl32, ccsA-ndhD and rps15-ycf1 showed high level of intrageneric variation. The phylogeny revealed that all the Veronica species formed a maximum supported (bootstrap value = 100) clade, and V. arvensis was sister to the clade formed by V. persica Poir. 1808 and V. polita Fries 1819 within the genus Veronica ().
Figure 2. Plastid genome map of V. arvensis. The genes inside and outside of the circle are transcribed in the clockwise and counterclockwise directions, respectively.
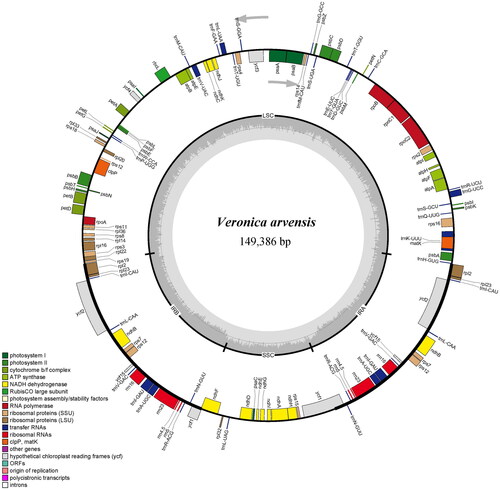
Figure 3. Comparative analysis of the nucleotide variability (Pi) values among seven Veronica species.
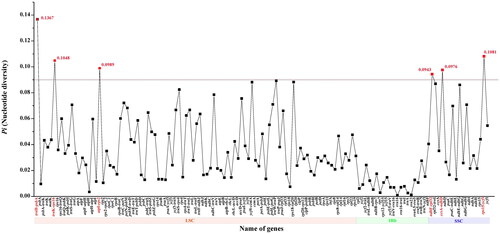
Figure 4. Phylogenetic tree of Plantaginaceae inferred using maximum likelihood (ML) based on complete plastome sequences. Numbers above the branches represent bootstrap values from ML analyses.
In the last ten years, high-throughput sequencing technologies have made it easier to obtain genomic data for any plant species, regardless of model organism status (Dodsworth Citation2015). The term ‘genome skimming’ introduced by Straub et al. (Citation2012), is a method of reducing sampling of low-coverage genome data by selective assembly of high-copy portions (plastome, mitogenome and repetitive elements). In many previous studies, complete plastomes have been successfully assembled using genome skimming data and subsequently used for phylogenetic reconstruction (Malé et al. Citation2015; Liu et al. Citation2017). Here, we assembled and reported the plastome of V. arvensis for the first time based on genome skimming data, the genome shared a common quadripartite structure similar to the majority of other angiosperms. Among the seven representative Veronica species, V. arvensis exhibited the smallest genome size and Veronica nakaiana had the largest one. The genome size variations of the plastomes were purely ascribed to the length differences of intergenic and intronic regions. Comparative plastome analysis showed that ycf15, which has been paid great attention to its function in previous studies (Raubeson et al. Citation2007; Shi et al. Citation2013), had been pseudogenized in V. persica and V. polita. The phylogeny inference reconstructed from complete plastomes revealed all the Veronica species formed a monophyletic clade and the branches were supported with higher bootstrap values than the results based on plastid, nuclear ribosomal and nuclear low-copy DNA (Albach and Meudt Citation2010), which proved that complete plastome sequences were more effective for the phylogenetic reconstruction of Veronica. Furthermore, six plastid hotspot regions identified among Veronica species will be useful in future studies reconstructing the phylogenetics relationships and characterizing the population genetics of this genus.
Author contributions
HW, HBL and WMH designed the study. QL conducted the field sampling. QL, ZMJ and CX produced the data. HBL, WMH, XBZ analyzed the data. HBL and WMH wrote the manuscript. HW, ZMJ and CX revised the manuscript. All authors approved the final manuscript.
Acknowledgments
We sincerely thank Hao Si for submitting the raw data to the GenBank database.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/under the accession no. ON461915. The associated Bio-Project, SRA, and Bio-Sample numbers of the raw sequence data for assembling the plastome are PRJNA835916, SRR19134697, and SAMN28125189, respectively.
Additional information
Funding
References
- Albach DC, Meudt HM. 2010. Phylogeny of Veronica in the Southern and Northern Hemispheres based on plastid, nuclear ribosomal and nuclear low-copy DNA. Mol Phylogenet Evol. 54(2):457–471.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Baskin J, Baskin C. 1983. Germination ecology of Veronica arvensis. J Ecol. 71(1):57–68.
- Dodsworth S. 2015. Genome skimming for next-generation biodiversity analysis. Trends Plant Sci. 20(9):525–527.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):31.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359.
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25(11):1451–1452.
- Liu LX, Li R, Worth JRP, Li X, Li P, Cameron KM, Fu CX. 2017. The complete chloroplast genome of Chinese bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front Plant Sci. 8:968.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(Web Server issue):W575–W581.
- Malé PJ, Bardon L, Besnard G, Coissac E, Delsuc F, Engel J, Lhuillier E, Scotti-Saintagne C, Tinaut A, Chave J. 2015. Genome skimming by shotgun sequencing helps resolve the phylogeny of a pantropical tree family. Mol Ecol Resour. 14(5):966–975.
- Mazur M, Marcysiak K, Dunajska A, Gawlak M, Kałuski T. 2021. Taxonomic significance of seed morphology in Veronica L. (Plantaginaceae) species from Central Europe. Plants. 11(1):88.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE), 2010. IEEE; p. 1–8.
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7):1253–1256.
- Raubeson LA, Peery R, Chumley TW, Dziubek C, Fourcade HM, Boore JL, Jansen RK. 2007. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics. 8:127–174.
- Shi C, Liu Y, Huang H, Xia EH, Zhang HB, Gao LZ. 2013. Contradiction between plastid gene transcription and function due to complex posttranscriptional splicing: an exemplary study of ycf15 function and evolution in angiosperms. PLoS One. 8(3):e59620.
- Straub SC, Parks M, Weitemier K, Fishbein M, Cronn RC, Liston A. 2012. Navigating the tip of the genomic iceberg: next-generation sequencing for plant systematics. Am J Bot. 99(2):349–364.
- Zhao Y. 1963. Experimental report on antimalarial effect of Veronica arvensis. J Tradit Chinese Med. 8:23. (In Chinese)
