Abstract
Arhopalus oberthuri is a pest which spreads in China, Laos, Japan and some other countries in Asia. The complete mitochondrial genome of A. oberthuri is 15,854 bp in length with 32.1% GC content, including 38.2% A, 20.4% C, 11.7% G, 29.7% T. There are 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNA) and two ribosomal RNA genes (rRNA) encoded in the genome. The graph of phylogenetic analysis gives the information that Arhopalus oberthuri is closer to Arhopalus unicolor. This study provided a scientific basis for the population genetics, phylogeny, and molecular taxonomy of A. oberthuri.
Arhopalus oberthuri Hua, 2002 belongs to Aseminae, a small family of beetles in the family Cerambycidae. The head of adult A. oberthuri is nearly circle-shaped with random dots. Two thirds of its mandibular is bare on the inside while rest of it is covered with pile on the outside (Peng Citation2019). They usually do not choose to breed on trees without bark. Most of the larvae stay in the underground part of the trees (Peng Citation2019). Arhopalus oberthuri can be found in Guangxi, Yunnan, Taiwan, Tibet and Fujian province in China. However, no reports about the genetic evolution of A. oberthuri have been published until now. Therefore, we reported the whole mitochondrial genome of A. oberthuri and built the maximum-likelihood tree inference method in search of the phylogeny relationship in this study.
The A. oberthuri Hua, 2002 adults were collected from Hongwei, Fujian Province, China (118°57′21″E, 26°9′19″N) by the traps with sexual attachments. We distinguished the specimens by the shape of genitals. The paramere of male genital takes up one-fourth of phallobase and the ventrolateral edge of copulatory opening isn’t blunt. The genital of female is long and thin and the edge of it is spindle-shaped, which makes them distinctive (Peng Citation2019). The specimens were deposited at the Fujian Agriculture and Forestry University (URL: https://lxy.fafu.edu.cn, contact person: Yinghua Tong and email: [email protected]) under the voucher number TN-202102. The total genomic DNA was extracted from one adult using TruSeq DNA sample Preparation kit (Vanzyme, CHN) and then purified by QIAquick Gel Extraction kit (Qiagen, GER). As for the determination of DNA quality and concentration, we chose NanoDrop 2000 (Thermo Fisher Scientific, USA). DNA sequencing was carried out by Illumina Hiseq 2500 (Illumina, USA). A total of 813,390 clean reads were obtained from 41,160,936 raw reads after filtration. MitoZ (Meng Citation2019) and metaSPAdes (Nurk et al. Citation2017) were used to assemble the clean reads. Then we used MITOs web server (Matthias et al. Citation2013) to annotate the assembly sequence and chose tRNAscan software (Lowe and Eddy Citation1997) to predict tRNA genes. The sequence of A. oberthuri has been submitted to NCBI GenBank with accession number OM161964.
The complete mitochondrial genome of Arhopalus oberthuri is 15,854 bp in length with 38.2% A, 29.7% T, 20.4% C and 11.7% G while the GC content is 32.1%. The genome has 13 protein-coding genes (PCGs), 22 tRNA genes, and two rRNA genes. The complete length of PCGs is 11,088 bp and 3695 amino acids are encoded. Among all the PCGs, only one PCG (cox2) starts with codon ATA, 7 PCGs (ND2, cox1, atp8, ND3, ND5, ND6, ND1) start with codon ATT and 5 PCGs start with codon ATG (atp6, cox3, ND4, ND4L, CYTB). As for the stop codon, there are 3 PCGs (atp8, ND3, ND1) stop with codon TAG and rest of them (ND2, cox1, ND5, ND6, cox2, atp6, cox3, ND4, ND4L, CYTB) stop with codon TAA.
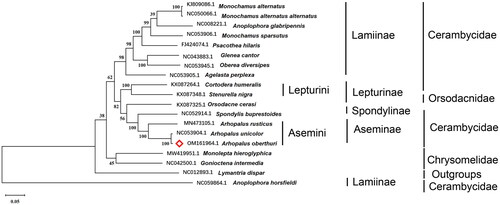
In order to confirm the phylogenetic position of A. oberthuri in Aseminae, the evolution tree was set with 18 relative species by MEGA 7.0 using Maximum Likelihood tree model with 1000 bootstrap replicates (Hall Citation2013). The method used orthofinder to analysis the rotholog genes among specimens. It was conducted using nucleotide sequences and 13 PCGs were used. The phylogenetic tree gives the information that A. oberthuri is closer to Arhopalus unicolor among these species (). All 18 relative species except Lymantria dispar are under the order Coleoptera and superfamily Chrysomeloidea. Also, Arhopalus rusticus and Arhopalus unicolor both belong to Aseminae. Since A. oberthuri is in the same subbranch as Arhopalus rusticus and Arhopalus unicolor, we can confirm that A. oberthuri is in Aseminae. The total mitochondrial genome of A. oberthuri will offer significant information which not only enriches the knowledge of the species of Aseminae but also helps us to study the evolution of Aseminae.
Ethical approval and consent form
The research, including the collection of animal materials, was carried out in accordance with guidelines provided by the authors’ institutions and national or international regulations.
Authors contribution
Xufei Pan and Yinghua Tong were involved in the conception and design; Xiaohong Han, Yunfei Liu and Yukun Zhu contributed the sample collection; Yunfei Liu, Shi Chen, Zengzeng Shi and Songqing Wu performed the analysis and interpretation of the data; Xufei Pan, Yukun Zhu and Shi Chen contributed the drafting of the paper; Xufei Pan and Yinghua Tong revised it critically for intellectual content. All authors were involved in the final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the assession No. OM161964. The associated BioProject, Bio-Sample and SRA numbers are PRJNA796479, SAMN24864354, SRR17575978, respectively.
Additional information
Funding
References
- Hall BG. 2013. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 30(5):1229–1235.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Matthias B, Alexander D, Frank J, Fabian E, Catherine F, Guido F, Joern P, Martin M, Peter FS. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Peng CL. 2019. Taxonomic and phylogeny of Chinese Spondylinae (Coleoptera, Cerambycidae). Chongqing: Southwest University.