Abstract
Schizothorax eurystomus, Kessler 1872 is a unique economic fish in Xinjiang, China that is rarely seen in the market. Next-generation sequencing (NGS) was used to determine the complete mitochondrial genome of S. eurystomus collected from the Yarkand River in Xinjiang. The results showed that the mitochondrial genome is a circular, 16,488-bp-long nucleotide with the typical vertebrate genome structure of 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and a control region. The termination-associated sequence (TAS), central conserved sequence block (CSB), and conserved sequence block were detected in the control region. Phylogenetic analysis placed S. eurystomus in a fully supported clade with S. biddulphi, and that clade was sister to S. yunnanensis. To our knowledge, this is the first study on the complete mitochondrial genome of S. eurystomus from the Yarkand River in Xinjiang, and it provides baseline genetic information for future studies.
Schizothorax eurystomus, Kessler 1872, also known as the wide-mouth hip scale fish, is a cold-water white fish that belongs to order Cypriniformes, family Cyprinidae, and subfamily Schizothoracinae (Cao et al. Citation2019). The S. eurystomus body is lengthened and slightly laterally flattened, and its head is tapered; S. eurystomus characteristics are shown in . S. eurystomus used to be the main economic fish in the Tarim River and its tributaries and affiliated lakes (Cao et al. Citation2019). Recently, with the impact of hydropower development and human activities, the S. eurystomus resources are decreasing, and the Xinjiang autonomous region plans to list it as a protected fish (Nie et al. Citation2014).
Next-generation sequencing (NGS) has revolutionized the field of molecular biology because it is rapid and can generate large amounts of genomic data (Schuster Citation2008; Koboldt et al. Citation2013). Therefore, in this study, the complete mitochondrial genome of S. eurystomus was determined using NGS technology to provide insight into the population processes and evolutionary history of this species (Zhang and Xian Citation2015).
DNA was extracted from muscle tissue of S. eurystomus collected from Altash Station (76.29E, 37.59 N) of the Xinjiang Yarkand River in September 2021. A specimen was deposited at the CAS Key Laboratory of Marine Ecology and Environmental Sciences, Institute of Oceanology, Chinese Academy of Sciences (Hui Jia, [email protected]) under voucher number L212. The Illumina NovaSeq sequencing platform (Illumina, San Diego, CA) was used to process sequences.
The high-quality second-generation genome sequence was assembled and analyzed using A5-miseq v20150522 (Coil et al. Citation2015) and SPAdes version 3.9.0 (Bankevich et al. Citation2012). Pilon version 1.18 (Walker et al. Citation2014) was used to correct the results and obtain the final mitochondrial sequence. Annotation of the complete mitochondrial genome sequence was performed on the MITOS web server (http://mitos2.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013). After sequencing, the maximum likelihood (ML) method with K2P distances was used to construct the ML tree in MEGA10 with 1000 bootstrap replicates (Kumar et al. Citation2018).
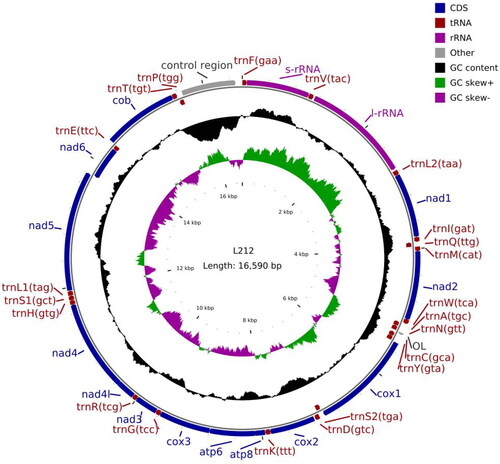
Similar to the size of other teleost mitogenomes, the complete mitochondrial genome of S. eurystomus was 16,488-bp long (GenBank accession ON920824). The circular map of the complete S. eurystomus mitochondrial genome is shown in . The complete mitochondrial genome contained 13 protein-coding genes, 2 ribosomal RNA (rRNA) genes (12S rRNA and 16S rRNA), 22 transfer RNA (tRNA) genes, and a control region, similar to other vertebrates (Miya et al. Citation2001).
Among the 13 protein-coding genes, ATP6 and ATP8 overlapped by 7 nucleotides, and NAD4 and NAD4L shared 7 nucleotides. The NAD5 and NAD6 genes overlapped by 4 nucleotides on the opposite strand. Most of the genes of S. eurystomus were encoded on the heavy-strand (H-strand), with only NAD6 and 8 tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, and tRNAPro) encoded on the light-strand (L-strand) (). The ATG codon initiated 12 of the 13 protein-coding genes (NAD1, NAD2, COII, ATP8, ATP6, COIII, NAD3, NAD4L, NAD4, NAD5, NAD6, and COB), and GTG was the initiation codon for COI (). The stop codon TAA terminated seven genes (NAD1, COI, AYP6, COIII, NAD4L, NAD5, and NAD6), TAG terminated three genes (NAD2, ATP8, and NAD3); the incomplete T– terminated the COII, NAD4, and COB genes by post-transcriptional polyadenylation (Ojala et al. Citation1981).
Table 1. Analysis of mitochondrial geneome characteristics.
The 12S and 16S rRNA genes of S. eurystomus were 956- and 1630-bp long, respectively. They were located between tRNAPhe and tRNALeu, and were separated by tRNAVal, similar to other vertebrates (Zhang et al. Citation2016). The 22 tRNA genes were scattered throughout the genome, ranging 67–76 bp and folded into a cloverleaf secondary structure with normal base shedding. The major non-coding region in S. eurystomus was located between tRNAPro and tRNAPhe, and was 821-bp long. The termination-associated sequence (TAS), central conserved sequence block (CSB), and CSB were detected in the control region and were similar to those of most bony fishes (Jia et al. Citation2021).
One complete mitogenome of S. eurystomus has been deposited in GenBank (https://www.ncbi.nlm.nih.gov/nuccore/KY436758.1), however, there is no paper published for the description in details on it. In this study, we collected and identified five specimens, and then used NGS to obtain their mitochondrial genome. The complete sequences were used for phylogenetic analysis and the phylogenetic results revealed that a previously collected specimen (KY436758.1) was genetically distant from other related species; therefore, the complete mitochondrial genome of S. eurystomus needed to be updated. Phylogenetic analysis of the complete mitochondrial genome of S. eurystomus revealed that it belonged to a clade with S. biddulphi, and they are sister to S. yunnanensis (). This study is the first to phylogenetically analyze S. eurystomus in detail and provides a scientific basis for future molecular systematic and phylogenetic studies of bony fishes in Cyprinidae.
Ethical approval
This study did not involve ethical approval, and the field investigation was supported by the Xinjiang Fisheries Research Institute, Urumqi 830000, China.
Author contributions
Conceptualization, J.N., H.L., J.H., T.Z., H.Z.; methodology, J.N., H.Z.; software, H.L., J.H., T.Z.; formal analysis, J.N., H.L., J.H.; investigation, J.N., H.L., J.H.; resources, H.Z.; data curation, J.H., T.Z.; writing, J.N., H.L., J.H., T.Z., H.Z.; supervision, H.Z. All authors have read the published version of the manuscript and agreed to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at nucleotide database, https://www.ncbi.nlm.nih.gov/nuccore/ON920824, Associated BioProject, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA855635, BioSample accession number at https://www.ncbi.nlm.nih.gov/biosample/ SAMN29499325 and Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra/SRR19976742.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cao XQ, Li YH, Wei JNZ, Yang Z. 2019. Relationship of body mass, body length, and fatness of Schizothorax eurystomus. Acat Agric Boreali-Occidentalis Sin. 28(08):1380–1386.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589.
- Jia H, Xu H, Xian WW, Li YQ, Zhang H. 2021. The complete mitochondrial genome of the spiny red gurnard Chelidonichthys spinosus McClelland, 1844 (Scorpaeniformes: triglidae). Mitochondrial DNA B Resour. 6(3):980–982.
- Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. 2013. The next-generation sequencing revolution and its impact on genomics. Cell. 155(1):27–38.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Miya M, Kawaguchi A, Nishida M. 2001. Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol. 18(11):1993–2009.
- Nie ZL, Wei J, Ma ZH, Zhang L, Song W, Wang WM, Zhang J. 2014. Morphological variations of Schizothoracinae species in the Muzhati River. J Appl Ichthyol. 30(2):359–365.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290(5806):470–474.
- Schuster SC. 2008. Next-generation sequencing transforms today’s biology. Nat Methods. 5(1):16–18.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9(11):e112963.
- Zhang H, Wang W, Xian WW. 2016. The complete mitochondrial genome of Anguilla japonica (Anguilliformes, Anguillidae) collected from Yangtze estuary and the phylogenetic relationship in genus Anguilla. Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4421–4422.
- Zhang H, Xian WW. 2015. The complete mitochondrial genome of the larvae Japanese anchovy Engraulis japonicus (Clupeiformes, Engraulidae). Mitochondrial DNA. 26(6):935–936.