Abstract
‘Biasong’ (Citrus micrantha), native to the Southern Philippine Islands, is an important cultivated fruit tree but has no published plastome sequence. We assembled and characterized the C. micrantha chloroplast genome from accession in the germplasm collection of the Institute of Crop Science, University of the Philippines Los Baños. The complete chloroplast (cp) genome sequence is 159,928-bp long with 128 coding genes comprising 83 mRNA genes, 37 tRNA genes, and eight rRNA genes. Out of the mRNA genes, 45 genes code for photosynthesis, 30 genes code for self-replication, and five genes code for other functions. A phylogenetic analysis of the assembled genome, along with 24 Citrus species and three other Rutaceae species, identified Citrus aurantiifolia as its closest relative with available complete cpDNA sequence.
Citrus micrantha Wester Citation1915 (Rutaceae), often called ‘biasong’ in the Philippines, is a wild species from the subgenus Papeda (Nicolosi Citation2007). It is utilized by the natives as a hair wash but not consumed as food according to Wester (Citation1915); however, it is used as a condiment in the southern part of the country, i.e. Northern Mindanao and Central Visayas. It can also be an important source of leaf essential oils (Baccati et al. Citation2021). Together with C. reticulata Blanco (mandarins), C. maxima Burm. Merrill (pummelos), and C. medica L. (citrons), these citrus species are known to be the foundation from which most cultivated citrus descended through interspecific hybridization (Nicolosi Citation2007; Curk et al. Citation2016). C. micrantha and var. microcarpa are of great interest to the scientific community as they are hypothesized to be the most primitive members of the subgenus. Even with its vital role in the evolution of Citrus and its economic significance, there is still no reported chloroplast (cp) genome sequence. Therefore, we assembled, annotated, and characterized the complete plastome of C. micrantha.
We collected fresh leaves of C. micrantha conserved in the field gene bank at the Crop Breeding and Genetic Resources Division, Institute of Crop Science, University of the Philippines Los Baños, Laguna. Its type locality is at Cagayan de Oro City (8° 29′ 7.3212″, 124° 43′ 58.9758″). The voucher specimen (ICROPS 1372) was consequently deposited in the Philippine Herbarium of Cultivated Plants of the Institute of Crop Science, University of the Philippines Los Baños, Laguna, Philippines (https://cafs.uplb.edu.ph/icrops/, Renerio P. Gentallan Jr., [email protected]). The genomic DNA from the fresh samples was extracted using the slightly modified CTAB method (Doyle and Doyle Citation1987) and was sent to NovogeneAIT Genomics Singapore PTE LTD (Singapore), for sequencing using HiSeq-PE150 platform (Illumina Inc., San Diego, CA). The 150-bp pair-end raw reads were produced and were sequentially filtered to generate 25,653,706 cleaned reads. The cp genome was assembled using the GetOrganelle v1.7.5+ (Jin et al. Citation2020), generating a circular genome. The circularized genome was then annotated and mapped using CPGAVAS2 (Shi et al. Citation2019) and was visualized using OGDRAW (Greiner et al. Citation2019). The assembled cp genome sequence was submitted to GenBank with the accession number ON597621 of the National Center for Biotechnology Information (NCBI). Accordingly, we characterized the morphology of the accession (ICROPS 1372) based on the notable morphological markers in the Citrus group described by Wester (Citation1915), for prospective revisions. This is also to verify that the genotype is closely associated with a certain reference specimen conserved in the GenBank and preserved in the herbarium. The plant, inflorescence, and fruits were photographed in situ and ex situ. Measured quantitative traits are reported as the mean ± standard deviation of the mean while qualitative traits were observed.
It is a shrub to tree (), with comparatively small flowers, white, in terminal cymes (). Its fruit is 49.46 ± 2.60 mm long, 33.48 ± 2.29 mm in transverse diameter, 24.8 ± 3.33 g in weight (), locules is 7–10 (), moderate olive green peel, pale yellow green flesh and pale yellow green placental hairs (), prolate shaped, rounded toward the base with bluntly pointed apices (); peel, placenta and exocarp thickness 0.86 ± 0.24 mm, 2.23 ± 0.93 mm, and 11.55 ± 1.88 mm, respectively; exocarp 10.79 ± 1.50 g; placenta 6.10 ± 0.77 g in weight; pulp is 9°Brix and 12°Brix at economic and physiological maturity, respectively. Many seeds (13–27) weigh 0.91 ± 0.28 g that are flat and pointed. The observed characteristics of the accession (ICROPS 1372) used for the assembly are within the range of the description delimited for C. micrantha, having small flowers with fewer stamens than any other species, oblong-obovate and few-loculed (6–10) fruits (Wester Citation1915).
Figure 1. Morphological characteristics of Citrus micrantha (ICROPS 1372), the specimen used for the chloroplast genome sequencing. C. micrantha plant (a), inflorescence (b), fruit at economic maturity (c), and physiological maturity (d).

Figure 2. Morphological characteristics of C. micrantha fruit (ICROPS 1372), the specimen used for the cp genome sequencing longitudinal cross-section (a), transverse cross-section (b), and fruit at economic maturity (c).

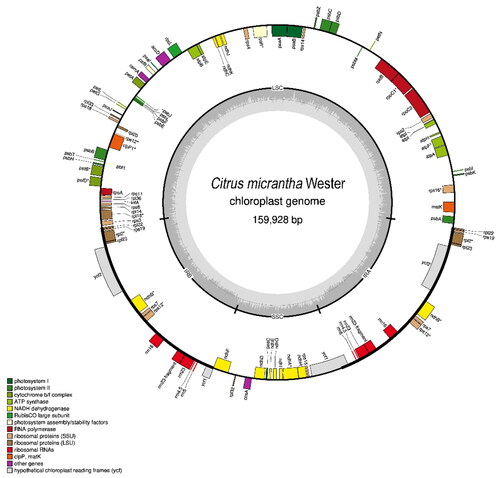
The assembled complete cp genome of C. micrantha was 159,928 bp in length. It consists of four distinct regions such as a pair of inverted repeats (IRs) regions (26,991 bp each), a short single-copy region (SSC) (18,768 bp) and a long single-copy (LSC) (87,178 bp) region. A total of 128 genes comprising 83 mRNA genes, 37 tRNA genes, and eight rRNA genes were annotated. Among these are 45 genes for photosynthesis, 30 genes for self-replication, three unknown conserved open reading frames (ycf15 x 2 and ycf4), and five other genes (accD, ccsA, cemA, clpP, and matK). The total GC content of the genome is 38.4% which includes 33.4% A, 33.4% T, 16.0% G, and 17.1% C. This is similar to C. aurantiifolia (KJ865401.1); however, the C. micrantha plastome is 35 bp longer (Su et al. Citation2014). C. micrantha has longer LSC and SSC regions as compared to C. aurantiifolia ().
Figure 3. Chloroplast gene map of C. micrantha genome. The inside and outside genes of the circle are transcribed in clockwise and counterclockwise directions, respectively. Different colors indicate genes belonging to different functional categories.
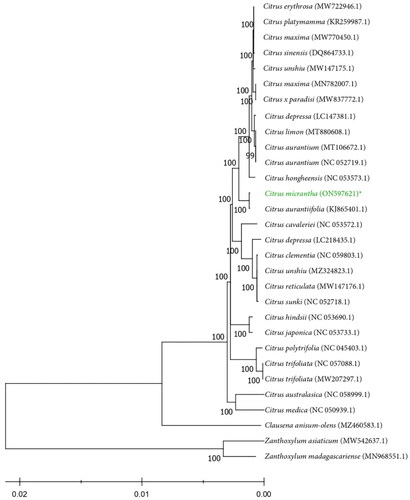
The complete cp DNA sequences of 24 other Citrus species and three species from other Rutaceae genera were downloaded from the NCBI database. The assembled genome sequence of C. micrantha along with the downloaded sequences, was aligned using MAFFT (Katoh and Standley Citation2013) and phylogenetic analysis was performed using MEGA-X (Kumar et al. Citation2018). These processes generated a maximum-likelihood (ML) tree with the General Time Reversible, Gamma-Invariant (GTR + G + I) as the model (Nei and Kumar Citation2000) with 1000 bootstrap replicates. The phylogenetic analysis showed that C. micrantha is closely related to C. aurantiifolia (Mexican lime) (). Biasong is believed to be the progenitor of the Mexican lime (Khan and Grosser, Citation2004). This could support the close relationship in the nomenclature history between the two Citrus species. C. aurantiifolia is purported to be a hybrid of C. micrantha Wester and sour orange (C. aurantium).
Ethics statement
The collection of plant material was carried out in accordance with guidelines provided by the authors’ institution (Institute of Crop Science, College of Agriculture and Food Science, University of the Philippines Los Baños).
Author contributions
All authors agree to be accountable for all aspects of the work. Roselle E. Madayag conceptualized the study, performed the experiments, analyzed the data, and wrote the paper; Renerio P. Gentallan, Kristine Joyce Quiñones, Michael Cedric Bartolome, Juan Rodrigo Vera Cruz, and Emmanuel Bonifacio S. Timog planted, collected, and prepared the germplasm, pressed the herbarium samples, reviewed literature, performed experiments, edited the paper; Teresita H. Borromeo and Leah Endonela helped conceptualized the study, validated the design of the experiment and the data presented, reviewed drafts of the paper.
Acknowledgements
The authors would also like to thank Ronil Beliber and Arvin Medrano for cultivating the germplasm, Eddelaine Joyce Bautista and Edna Mercado for processing the paperwork needed for the research project and Arcelle Mae S. Daayata for the photographs.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. ON597621. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA867485, SRR20981937, and SAMN30201906, respectively.
References
- Baccati C, Gibernau M, Paoli M, Ollitrault P, Tomi F, Luro F. 2021. Chemical variability of peel and leaf essential oils in the citrus subgenus Papeda (Swingle) and few relatives. Plants. 10(6):1117.
- Curk F, Ollitrault F, Garcia-Lor A, Luro F, Navarro L, Ollitrault P. 2016. Phylogenetic origin of limes and lemons revealed by cytoplasmic and nuclear markers. Ann Bot. 117(4):565–583.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Khan IA, Grosser JW. 2004. Regeneration and characterization of somatic hybrid plants of Citrus sinensis (sweet orange) and Citrus micrantha, a progenitor species of lime. Euphytica. 137(2):271–278.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. New York: Oxford University Press.
- Nicolosi E. 2007. Origin and taxonomy. In: Khan IA, editor. Citrus genetics, breeding and biotechnology. Oxfordshire (UK): CAB International; p. 19–44.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Su H-J, Hogenhout SA, Al-Sadi AM, Kuo C-H. 2014. Complete chloroplast genome sequence of omani lime (Citrus aurantiifolia) and comparative analysis within the rosids. PLOS One. 9(11):e113049.
- Wester H-J. 1915. Citrus fruits in the Philippines. Philippine Agric Rev. 8(1):20–21.