Abstract
The complete nucleotide sequence of the mitochondrial (mt) genome of the demersal zebra seabream Diplodus cervinus (Lowe, 1838) was determined for the first time. The double stranded circular molecule is 16,559 base pairs (bp) in length and encodes for the typical 37 metazoan mitochondrial genes, and 2 non-coding regions (D-loop and L-origin). The gene arrangement of the D. cervinus mt genome follows the usual one for fishes. The nucleotide sequences of the mt protein coding and ribosomal genes of D. cervinus mt genome were aligned with orthologous sequences from representatives of the Sparidae family and phylogenetic relationships were inferred. Maximum likelihood analyses placed D. cervinus as a sister species of Diplodus sargus (Linnaeus, 1758).
The zebra seabream Diplodus cervinus (Lowe, 1838) is a gregarious demersal marine fish of the family Sparidae Rafinesque, 1818, usually living in groups of 4–5 individuals over rocky bottoms up to 80 m depth, although it can be also found on muddy bottoms up to 300 m (Bauchot and Hureau Citation1986; Pajuelo, Lorenzo, and Domı́nguez-Seoane Citation2003). This thermophilic species is distributed in the eastern Atlantic Ocean from the Bay of Biscay to Cape Verde Islands, from Angola to South Africa, and around Madeira and Canary Islands, but it also occurs in the Mediterranean Sea, where it is recently widening its distribution (Bauchot and Hureau Citation1986; Pajuelo, Lorenzo, and Domı́nguez-Seoane Citation2003; Tiralongo et al. Citation2020). It reaches ∼55 cm in length and 2.7 kg in weight, and it is a species of interest in small scale fisheries throughout its range of distribution, with scattered attempts to rear it using aquaculture techniques (Bauchot and Hureau Citation1986; IGFA Citation2001). This species, together with other ones of the genus Diplodus, represents a candidate with great potential for aquaculture, due to its market price and good adaptability to farming environment (Coutinho et al. Citation2016). In some coastal areas, like those of Canary Islands, this demersal species covers a relevant ecological role (Pajuelo, Lorenzo, Domı́nguez, et al. Citation2003).
The D. cervinus specimen analyzed in this study was meant for sale as seafood to the consumers. The specimen was caught by local fisherman and it was collected for research as a dead specimen from the fisherman’s market (36.7406 N, 15.1193E), where it was supplied directly from local fishermen. Thus, it did not undergo any manipulation or experimentation in the laboratory. Its usage for scientific purposes is not included in the Article 2 of the Italian Legislative Decree n. 26 of 4 March 2014, national transposition of the European Directive 2010/63/UE. Complete mt genome sequence of D. cervinus and its annotation is presented here for the first time. The specimen was caught with trammel nets (1.5 mt depth) on a rocky bottom off Marzamemi (Syracuse, Sicily, Italy; Ionian Sea, Mediterranean Sea, ∼36.7480 N, 15.1129E) on 18 April 2020. It was morphologically identified based on species-specific diagnostic characters and subsequently deposited in the Darwin Dohrn Museum of the Stazione Zoologica Anton Dohrn of Naples (http://193.205.231.138/ZooColl/HTML/index.php, curator Andrea Travaglini, [email protected]) with the code number SZN-OST-0003. Total DNA was extracted from 25 mg of dorsal muscle tissue following Mascolo, Ceruso, Sordino, et al. (Citation2019) methodology. The assembled and annotated mitogenome was obtained by high-throughput sequencing of enriched mitochondrial DNA with Illumina NextSeq 550 System (Illumina, San Diego, CA, USA). The obtained sequences were assembled using MegaHit (Li et al. Citation2015) through the Galaxy server at https://usegalaxy.eu/ (Afgan et al. Citation2018). The final contig obtained was annotated by the MitoFish server (Iwasaki et al. Citation2013) and subsequently checked manually.
The D. cervinus mitogenome was 16,559 bp long. The overall nucleotide composition was: 27.58% A, 29.30% C, 26.56% T, and 16.56% G, being similar to other Sparidae mitogenome data (Ceruso et al. Citation2018a, Citation2018b, Citation2020; Mascolo et al. Citation2018a, Citation2018b; Mascolo, Ceruso, Chirollo, et al. Citation2019; Caputi et al. Citation2021). As is the case for most metazoans (Boore Citation1999), the mtDNA encoded for 13 protein-coding genes, 22 tRNAs, and 2 rRNAs. Also, two non-coding regions (D-loop and L-origin) were present, in agreement with fish mitochondrial genomes (Satoh et al. Citation2016). The heavy strand of the mt genome encoded for 12 protein-coding genes, the majority of the tRNA genes, and the 2 ribosomal genes (12S and 16S). The NADH dehydrogenase subunit 6 (nad6) gene and the trnA, trnN, trnC, trnY, trnE, and trnC were encoded by the light strand. The mt genome organization followed those previously described (see Ceruso et al. Citation2020; Fietz et al. Citation2020; Caputi et al. Citation2021).
All protein-coding genes started with the codon ATG except for the subunit 1 of the cytochrome oxidase (cox1) that started with GTG. Some genes had complete stop codons (TAA in nad4L and nad5; TAG in subunit 8 of the ATP synthase (atp8), nad1 and nad6; AGG in cox1), while other genes ended with a single T (cox2, cob, nad3, and nad4) or TA (atp6, cox3, and nad2), which presumably becomes functional by subsequent polyadenylation of the transcribed messenger RNA (Ojala et al. Citation1981).
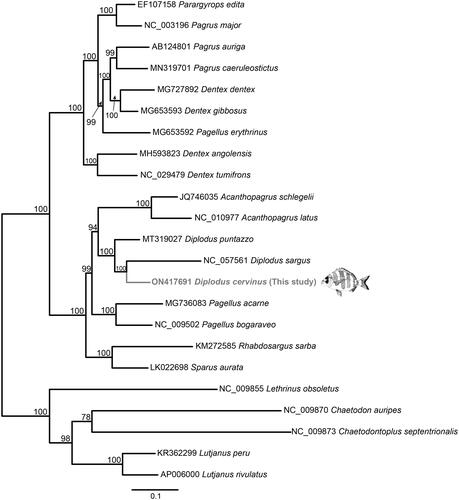
A maximum likelihood (ML) analysis was implemented to elucidate the phylogenetic position of D. cervinus. It was performed in RAxML-NG (Kozlov et al. Citation2019). The resultant phylogeny () placed D. cervinus as sister species of D. sargus (Linnaeus, 1758) with maximum support, and both species as sister group of Diplodus puntazzo (Walbaum, 1792), again with maximum support. So, all three latter species formed a monophyletic group that corresponded to a clade including the species of the genus Diplodus Rafinesque, 1810. This clade was placed in the topology as a sister group of a clade including the species of the genus Acanthopagrus Peters, 1855, with a ML support of 94. This last clade was recovered as sister group of Pagellus acarne (Risso, 1827) and Pagellus bogaraveo (Brünnich, 1768), similar to previous studies (Ceruso et al. Citation2020; Caputi et al. Citation2021).
Figure 1. Phylogenetic relationships in the family Sparidae based on the complete mt genome sequences available in GenBank and that of Diplodus cervinus reported here (Acanthopagrus latus NC_010977; Acanthopagrus schlegelii JQ746035; Dentex angolensis MH593823; Dentex MG727892; Dentex gibbosus MG653593; Dentex tumifrons NC_029479; Diplodus puntazzo MT319027; Diplodus sargus NC_057561; Pagellus acarne MG736083; Pagellus bogaraveo NC_009502; Pagellus erythrinus MG653592; Pagrus auriga AB124801; Pagrus caeruleostictus MN319701; Pagrus major NC_003196; Parargyrops edita EF107158; Rhabdosargus sarba KM272585; Sparus aurata LK022698). Five outgroup species (Lutjanus peru KR362299, Lutjanus rivulatus AP006000, Lethrinus obsoletus NC_009855, Chaetodontoplus septentrionalis NC009873, and Chaetodon auripes NC_009870) were selected. Maximum likelihood method was used with an automatic bootstrapping cutoff of 0.01.
Ethical approval
This study has been reviewed by the Ethical Animal Care and Use Committee of the University of Naples Federico II and received institutional approval (Notice No. PG/2022/0093423, July 27th 2022).
Author contributions
FC, TP and PS conceived and designed the project. VT, RMS, AL, FT, IV and MC performed labwork. DO, LC and FT performed data analyses and interpretation. DO drafted the manuscript. DO, TP, LC, VT, RMS, AL, FT, IV, FB, and PS revised the manuscript for intellectual content and were involved for final approval of the version to be published. All the authors agreed to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. ON417691. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA836136, SRR19136359, and SAMN28132046, respectively.
References
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Grüning BA, et al. 2018. The Galaxy platform for accessible, reproducible, and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46(W1):W537–W544.
- Bauchot ML, Hureau JC. 1986. Sparidae. In: Whitehead PJP, Bauchot ML, Hureau JC, Nielsen J, Tortonese E, editors. Fishes of the north-eastern Atlantic and the Mediterranean. Vol. 2. Paris: UNESCO; p. 883–907.
- Boore J. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Caputi L, Osca D, Ceruso M, Venuti I, Sepe RM, Anastasio A, D'Aniello S, Crocetta F, Pepe T, Sordino P. 2021. The complete mitochondrial genome of the white seabream Diplodus sargus (Perciformes: Sparidae) from the Tyrrhenian Sea. Mitochondrial DNA B Resour. 6(9):2581–2583.
- Ceruso M, Mascolo C, Lowe EK, Palma G, Anastasio A, Pepe T, Sordino P. 2018b. The complete mitochondrial genome of the common Pandora Pagellus erythrinus (Perciformes: Sparidae). Mitochondrial DNA B Resour. 3(2):624–625.
- Ceruso M, Mascolo C, Palma G, Anastasio A, Pepe T, Sordino P. 2018a. The complete mitochondrial genome of the common dentex, Dentex (Perciformes: Sparidae). Mitochondrial DNA B Resour. 3(1):391–392.
- Ceruso M, Venuti I, Osca D, Caputi L, Anastasio A, Crocetta F, Sordino P, Pepe T. 2020. The complete mitochondrial genome of the sharpsnout seabream Diplodus puntazzo (Perciformes: Sparidae). Mitochondrial DNA B Resour. 5(3):2379–2381.
- Coutinho F, Peres H, Castro C, Pérez-Jiménez A, Pousão-Ferreira P, Oliva-Teles A, Enes P. 2016. Metabolic responses to dietary protein/carbohydrate ratios in zebra sea bream (Diplodus cervinus, Lowe, 1838) juveniles. Fish Physiol Biochem. 42(1):343–352.
- Fietz K, Trofimenko E, Guerin PE, Arnal V, Torres-Oliv M, Lobréaux S, Pérez-Ruzafa A, Manel S, Puebla O. 2020. New genomic resources for three exploited Mediterranean fishes. Genomics. 112(6):4297–4303.
- IGFA. 2001. Database of IGFA angling records until 2001. Fort Lauderdale (FL): IGFA.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh T, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455. DOI: 10.1093/bioinformatics/btz305.
- Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 31(10):1674–1676.
- Mascolo C, Ceruso M, Chirollo C, Palma G, Anastasio A, Sordino P, Pepe T. 2019. The complete mitochondrial genome of the Angolan dentex angolensis (Perciformes: Sparidae). Mitochondrial DNA B Resour. 4(1):1245–1246.
- Mascolo C, Ceruso M, Palma G, Anastasio A, Sordino P, Pepe T. 2018a. The complete mitochondrial genome of the axillary seabream, Pagellus acarne (Perciformes: Sparidae). Mitochondrial DNA B Resour. 3(1):434–435.
- Mascolo C, Ceruso M, Palma G, Anastasio A, Sordino P, Pepe T. 2018b. The complete mitochondrial genome of the Pink dentex gibbosus (Perciformes: Sparidae). Mitochondrial DNA B Resour. 3(2):525–526.
- Mascolo C, Ceruso M, Sordino P, Palma G, Anastasio A, Pepe T. 2019. Comparison of mitochondrial DNA enrichment and sequencing methods from sparid fishes. Food Chem. 294:333–338.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290(5806):470–474.
- Pajuelo JG, Lorenzo JM, Domı́nguez-Seoane R. 2003. Age estimation and growth of the zebra seabream Diplodus cervinus (Lowe, 1838) on the Canary Islands shelf (Central-east Atlantic). Fish Res. 62(1):97–103.
- Pajuelo JG, Lorenzo JM, Domínguez R, Ramos A, Gregoire M. 2003. On the population ecology of the zebra seabream Diplodus cervinus (Lowe 1838) from the coasts of the Canarian archipelago. North West Africa. Environ. Biol. Fish. 67(4):407–416.
- Satoh TP, Miya M, Mabuchi K, Nishida M. 2016. Structure and variation of the mitochondrial genome of fishes. BMC Genom. 17(1):1–20.
- Tiralongo F, Crocetta F, Riginella E, Lillo AO, Tondo E, Macali A, Mancini E, Russo F, Coco S, Paolillo G, et al. 2020. Snapshot of rare, exotic and overlooked fish species in the Italian seas: a citizen science survey. J Sea Res. 164:101930.
