Abstract
The climbing plant Cynanchum rostellatum (Turcz.) Liede & Khanum is widely distributed throughout Korea and Northeast Asia as a member of the Apocynaceae family. Although this plant has a high value in medicinal and industrial purposes, genetic research on this plant is insufficient. This study announces the complete plastid genome (plastome) sequence of C. rostellatum with 663× mean coverage, which was assembled using 763 Mbp short-read data generated by the Illumina HiSeq X platform. The C. rostellatum plastome was 158,018 bp in length and displayed the typical quadripartite structure composed of the large single-copy (LSC) region (89,058 bp), the small single-copy (SSC) region (18,718 bp), and a pair of inverted repeat (IR) regions (25,116 bp). A total of 129 genes have been annotated, including 84 protein-coding genes, 37 transfer RNA genes, and eight ribosomal RNA genes. Phylogenetic analysis indicated the genus Cynanchum including 12 Cynanchum plastome sequences, was monophyletic and was located within the sub-family Asclepiadoideae. Two C. rostellatum plastomes, including the plastome assembled in this study, formed a subclade and were sister to the C. thesioides plastome, whereas the other C. rostellatum, which was previously reported one, was located within the clade of C. wilfordii and C. bungei.
Cynanchum rostellatum (Turcz.) Liede & Khanum (Citation2016) is a perennial plant belonging to the Apocynaceae family and Asclepiadoideae sub-family and has white milky sap and climbing vines (Khanum et al. Citation2016) (). The root of this species is widespread and has many buds for vegetative propagation. In addition, their seeds have feathers for propagation (Kim et al. Citation2014). This species previously belonged to the genus Metaplexis and was recognized as Metaplexis japonica (Thunb.) Makino (1903). However, the genus Metaplexis was recently merged with the genus Cynanchum based on molecular evidence of several barcoding regions, including plastid and nuclear regions (Khanum et al. Citation2016). The C. rostellatum has traditionally been used as a medicinal plant for erectile dysfunction (Wei et al. Citation2019). Additional functions such as antibacterial, antioxidant, and nerve cell protection effects have also been reported (Jamarkattel-Pandit and Kim Citation2019; Wei et al. Citation2019). Moreover, the feathers on the seeds can separate oil from water, increasing their industrial value (Wang et al. Citation2019). Although C. rostellatum is morphologically similar to other Cynanchum species with climbing vines, such as C. wilfordii (Kim et al. Citation2014). This species can be distinguished from other species by having an elongated stigma and purple furry petals in flower morphology (Nam and Chung Citation2018). However, when flowers fall, C. rostellatum is frequently misidentified as C. wilfordii, one of the most famous medicinal plants in Korea, especially during the harvesting season (Kim et al. Citation2014).
Figure 1. The specimen and morphology of C. rostellatum. (a) The specimen of C. rostellatum (voucher number: SNUA00057640). (b) The shape of flowers and leaves of C. rostellatum.
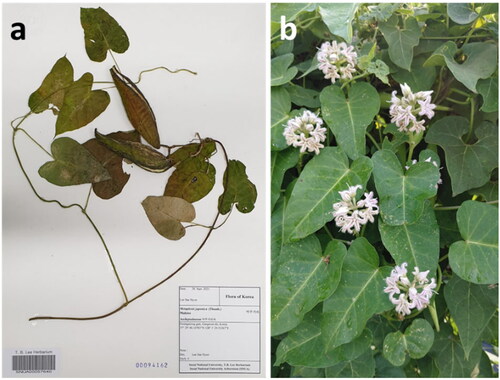
Therefore, molecular data of this species are still required for species authentication between C. rostellatum and other Cynanchum species. This study provides information regarding the plastome evolution and intra-species diversity of C. rostellatum, which is essential for further species authentication and evolutionary research.
The leaf material of C. rostellatum was collected from Hoengseong-gun, Kangwon Province, South Korea (37° 29′ 40.10783″N, 128° 1′ 29.53267″E), and the specimen was deposited in the T.B. Lee Herbarium of Seoul National University (http://arbor.snu.ac.kr/eng/, Sook-Hyang Kim, [email protected]) under the voucher number SNUA00057640. Genomic DNA was extracted from newly sprouted young leaves using the GeneAll Exgene plant midi kit (Geneall Biotechnology Ltd., Seoul, South Korea). Pair-end library construction was conducted using HiSeq X Reagent Kit (Illumina, San Diego, CA), and the insert size of 550 bp. Sequencing was conducted on the Illumina HiSeq X platform (Lab genomics LLC, Seongnam, South Korea). The total read length of raw data was 763,415,230 bp. The following information describes how the complete plastome was assembled using the dnaLCW method (Kim et al. Citation2015). De novo assembly was conducted with trimmed raw data using CLC Genomics Workbench (version 10.0.3, CLC Inc., Aarhus, Denmark). Among assembled contigs, plastome sequences were retrieved, ordered, and merged into a single sequence, using the C. wilfordii plastome sequence as the reference (Lee et al. Citation2022). Remained gaps were closed with SOAP Gapcloser (http://soap.genomics.org.cn). The average and minimum read mapping depth of assembled genome were 663.34× and 80×, respectively (Figure S1). Gene annotation of the plastome sequence was conducted using GeSeq (Tillich et al. Citation2017) and then manually curated. The plastome map was drawn using the CPGView program (http://www.1kmpg.cn/cpgview).
For phylogenetic analysis, a total of 38 plastome sequences were downloaded from the NCBI GenBank. Members of the subfamily Rauvolfioideae, Rhazya stricta (KJ123753), and Catharanthus roseus (NC 021423) were used as an outgroup. From 39 plastome sequences, 50 co-existing protein-coding genes were selected and merged from each plastome sequence. Sequences were aligned with the MAFFT (Katoh and Standley Citation2013) and trimmed using Gblocks (Talavera and Castresana Citation2007). The ModelFinder option of the IQ-TREE program figured out the best tree model, and the TVM + F+I + G4 substitution model was selected for the analysis (Nguyen et al. Citation2015). A maximum-likelihood (ML) tree was constructed using the IQ-TREE (Nguyen et al. Citation2015) with 1000 bootstrap replicates. Using in-house Python code, sequence variations were counted from the aligned sequences of three C. rostellatum.
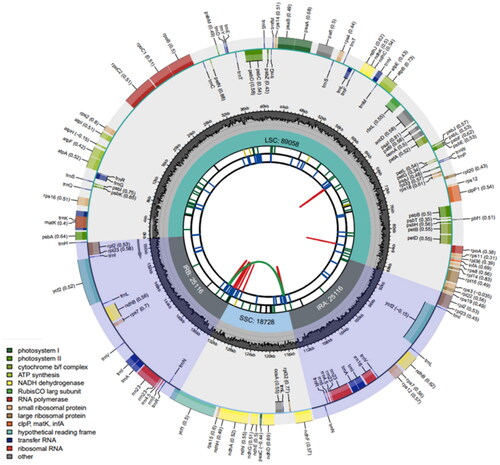
The plastome of C. rostellatum was 158,018 bp in length, organized into a quadripartite structure, including large single-copy (LSC) (89,058 bp), small single-copy (SSC) (18,728 bp), and inverted repeat (IR) (25,116 bp) regions. The GC contents of the LSC, SSC, and IR regions were 36.3%, 32.2%, and 43.1%, respectively. A total of 129 genes were annotated, consisting of 84 protein-coding genes, 37 tRNA genes, and eight rRNA genes ().
Figure 2. Plastome genome map of C. rostellatum. From the inner circle, the first circle depicts distributed repeats connected by red (forward direction) and green (reverse direction) arcs, respectively. The following circle displays tandem repeats denoted by short blue bars. The sequences of microsatellites are depicted as short green bars. The fourth circle displays the sizes of LCS, SSC, and IR. The fifth circle illustrates the distribution of GC contents along the plastome (dark grey: GC contents, light grey: background). The sixth circle displays the genes with colored boxes. The outer and inner colored boxes present transcribed clockwise and counterclockwise genes, respectively.
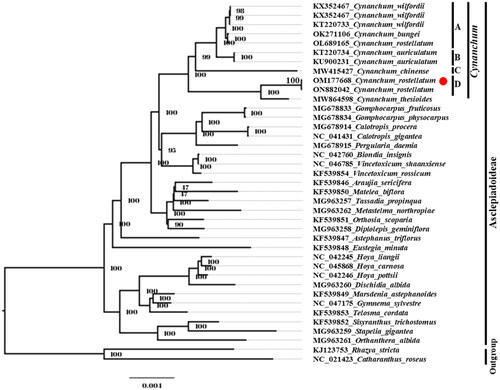
Our phylogenetic analysis indicated that the genus Cynanchum was monophyletic, and the clade of Cynanchum included four subclades (). Two C. rostellatum plastomes (OM177668 and ON882042) formed a subclade with the C. thesioides plastome (MW864598), and the subclade was sister to other Cynanchum plastomes. The C. chinense plastome was sister to the clade of C. wilfordii, C. bungei, C. auriculatum, and one C. rostellatum (OL689165). The plastomes of C. wilfordii and C. auriculatum formed independent subclades. However, one C. rostellatum plastome (OL689165) formed a subclade with the C. bungei plastome. The conflict positions among three C. rostellatum plastomes in the phylogeny were also supported by the sequence variation analysis. The result showed that 34 variations (16 SNPs and 18 InDels) were found between two plastomes, OM177668 and ON882042, whereas 4579 variations (3776 SNPs and 803 InDels) were found between OM177668 and OL689165. The great intra-specific diversity in C. rostellatum implies the necessity of further research for this species in terms of genetic diversity and taxonomic treatment. This study provides fundamental information for further studies on species authentication of C. rostellatum and the evolution of the Apocynaceae family.
Figure 3. Phylogenetic analysis of C. rostellatum with allied species. A phylogenetic tree was constructed using the maximum-likelihood (ML) method with 1000 bootstrap replicates and 50 common protein-coding gene sequences from the plastome. The red dot represents the assembled plastome sequence in this study. The clades of species are represented with black lines. GenBank accession numbers are as follows: C. wilfordii KX352467, C. wilfordii KT220733 (Park et al. Citation2016), C. bungei OK271106, C. rostellatum OL689165 (Pei et al. Citation2022), C. auriculatum KT220734 (Jang et al. Citation2016), C. auriculatum KU900231, C. chinense MW415427 (Chen and Zhang Citation2022), C. rostellatum ON882042, C. thesioides MW864598, Gomphocarpus fruticosus MG678833 (Fishbein et al. Citation2018), G. physocarpus MG678834 (Fishbein et al. Citation2018), Calotropis procera MG678914, C. gigantea NC_041431 (Fishbein et al. Citation2018), Pergularia daemia MG678915 (Fishbein et al. Citation2018), Biondia insignis NC_042760 (Guan and Zhang Citation2019), Vincetoxicum shaanxiense NC_046785 (Rao et al. Citation2018), V. rossicum KF539854 (Straub et al. Citation2013), Araujia sericifera KF539846 (Straub et al. Citation2013), Matelea biflora KF539850 (Straub et al. Citation2013), Tassadia propinqua MG963257 (Fishbein et al. Citation2018), Metastelma northropiae MG963262 (Fishbein et al. Citation2018), Orthosia scoparia KF539851 (Straub et al. Citation2013), Diplolepis geminiflora MG963258 (Fishbein et al. Citation2018), Astephanus triflorus KF539847 (Straub et al. Citation2013), Eustegia minuta MG678914 (Straub et al. Citation2013), Hoya liangii NC_042245 (Tan et al. Citation2018), H. carnosa NC_045868 (Wei et al. Citation2020), H. pottsii NC_042246 (Tan et al. Citation2018), Dischidia albida MG963260 (Fishbein et al. Citation2018), Marsdenia astephanoides KF539849 (Straub et al. Citation2013), Gymnema sylvestre NC_047175, Telosma cordata KF539853 (Straub et al. Citation2013), Sisyranthus trichostomus KF539852 (Straub et al. Citation2013), Stapelia gigantea MG963259 (Fishbein et al. Citation2018), Orthanthera albida MG963261 (Fishbein et al. Citation2018), Rhazya stricta KJ123753, and Catharanthus roseus NC_021423 (Ku et al. Citation2013).
Ethical approval
This study complies with Seoul National University’s Research Ethics Guidelines, relevant institutional, national, and international guidelines and legislation. Permission was not required for sample collection in this study. C. rostellatum is widespread in South Korea and is not listed as a threatened or endangered species.
Author contributions
S. H. L., W. J., E. b., and T.-J. Y. planned and designed this research. J. K. and S. H. L. assembled and annotated the plastome sequence. J. K., H. G., J. S. K., H. S., and J. Y. P. collected samples, made specimens, and extracted DNA. S. H. L., W. J., and E. b. wrote the manuscript, J. K., H. G., J. S. K., H. S., and J. Y. P. revised the manuscript.
Supplemental Material
Download MS Word (50.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data supporting this study’s findings are openly available in GenBank of NCBI (accession number: OM177668). The associated BioProject is PRJNA765723, and this research was conducted as a part of the genome analysis of Cynanchum species. SRA and Bio-sample numbers are SRR17333880 and SAMN24429174, respectively.
Additional information
Funding
References
- Chen G, Zhang X. 2022. The complete chloroplast genome of Chinese medicinal herb Cynanchum chinense R. Br. (Apocynaceae) and its phylogenetic position. Mitochondrial DNA Part B. 7(4):598–599.
- Fishbein M, Livshultz T, Straub SC, Simões AO, Boutte J, McDonnell A, Foote A. 2018. Evolution on the backbone: Apocynaceae phylogenomics and new perspectives on growth forms, flowers, and fruits. Am J Bot. 105(3):495–513.
- Fishbein M, Straub SC, Boutte J, Hansen K, Cronn RC, Liston A. 2018. Evolution at the tips: Asclepias phylogenomics and new perspectives on leaf surfaces. Am J Bot. 105(3):514–524.
- Guan M, Zhang R. 2019. The complete chloroplast genome of Biondia insignis Tsiang (Apocynaceae). Mitochondrial DNA Part B. 4(1):280–281.
- Jamarkattel-Pandit N, Kim H. 2019. Neuroprotective effect of Metaplexis japonica against in vitro ischemia model. J Health Allied Sci. 3(1):51–55.
- Jang W, Kim K-Y, Kim K, Lee S-C, Park H-S, Lee J, Seong RS, Shim YH, Sung SH, Yang T-J. 2016. The complete chloroplast genome sequence of Cynanchum auriculatum Royle ex Wight (Apocynaceae). Mitochondrial DNA Part A. 27(6):4549–4550.
- Kang S-J, Park H-S, Koo HJ, Park JY, Lee DY, Kang KB, Han SI, Sung SH, Yang T-J. 2018. The complete chloroplast genome sequence of Korean Lonicera japonica and intra-species diversity. Mitochondrial DNA Part B. 3(2):941–942.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Khanum R, Surveswaran S, Meve U, Liede-Schumann S. 2016. Cynanchum (Apocynaceae: Asclepiadoideae): a pantropical Asclepiadoid genus revisited. Taxon. 65(3):467–486.
- Kim MJ, Kim IJ, Choi SY, Han DH, Kim YH, Lim SC, Kim TJ, Nam SY, Song BH, Oh BU, et al. 2014. Comparison of Cynanchum wilfordii, C. auriculatum, Metaplexis japonica and Polygonum multiflorum by morphological characters. Korean J Med Crop Sci. 22(2):113–120.
- Kim K, Lee S-C, Lee J, Yu Y, Yang K, Choi B-S, Koh H-J, Waminal NE, Choi H-I, Kim N-H. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5(1):1–13.
- Ku C, Chung W-C, Chen L-L, Kuo C-H. 2013. The complete plastid genome sequence of madagascar periwinkle Catharanthus roseus (L.) G. Don: plastid genome evolution, molecular marker identification, and phylogenetic implications in asterids. PLOS One. 8(6):e68518.
- Lee SH, Kim J, Park H-S, Koo H, Waminal NE, Pellerin RJ, Shim H, Lee H-O, Kim E, Park JY, et al. 2022. Genome structure and diversity among Cynanchum wilfordii accessions. BMC Plant Biol. 22(1):4–14.
- Nam B-m, Chung GY. 2018. Taxonomic implications of floral morphology in the subfamily Asclepiadoideae (Apocynaceae s.l.) in Korea. Korean J Plant Taxon. 48(3):172–184.
- Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Park H-S, Kim K-Y, Kim K, Lee S-C, Lee J, Seong RS, Shim YH, Sung SH, Yang T-J. 2016. The complete chloroplast genome sequence of an important medicinal plant Cynanchum wilfordii (Maxim.) Hemsl. (Apocynaceae). Mitochondrial DNA Part A. 27(5):3747–3748.
- Pei L, Shu S, Ji B, Cui N. 2022. Complete sequence of Cynanchum rostellatum (Apocynaceae: Asclepiadoideae) chloroplast genome and its phylogenetic analysis. Mitochondrial DNA Part B. 7(7):1395–1397.
- Rao H, Wang X-J, Ma J-X, Li Q-E, Li G-L, Zou J-B. 2018. Characterization of the complete chloroplast genome of Biondia chinensis (Apocynaceae: Asclepiadoideae: Asclepiadeae), a rare and threatened liana endemic to China. Mitochondrial DNA Part B. 3(2):763–764.
- Straub SC, Cronn RC, Edwards C, Fishbein M, Liston A. 2013. Horizontal transfer of DNA from the mitochondrial to the plastid genome and its subsequent evolution in milkweeds (Apocynaceae). Genome Biol Evol. 5(10):1872–1885.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577.
- Tan X-H, Wang J-H, Zhao K-K, Zhu Z-X, Wang H-F. 2018. Complete plastome sequence of Hoya pottsii Traill and Hoya liangii Tsiang (Apocynaceae). Mitochondrial DNA Part B. 3(2):1176–1177.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq– versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang Z, Wang D, Wang M, Li W, Sui Q. 2019. Metaplexis japonica seed hair fiber: a member of natural hollow fibers and its characterization. Text Res J. 89(21–22):4363–4372.
- Wei L, Yang M, Huang L, Li JL. 2019. Antibacterial and antioxidant flavonoid derivatives from the fruits of Metaplexis japonica. Food Chem. 289:308–312.
- Wei X-F, Zeng S-J, Zhang G-Q, Tang G-D, Huang J-X. 2020. Complete plastome sequence of Hoya carnosa (L. f.) R. Br. (Apocynaceae). Mitochondrial DNA Part B. 5(1):522–523.
