Abstract
Primula calliantha subsp. bryophila (Balf. f. et Farrer) W.W. Smith and Forrest (1928) is a perennial alpine species with ornamental value. It is distributed in northwestern Yunnan and adjacent eastern Tibet of China, and northern Myanmar. Here, we sequenced and assembled complete plastid genome of P. calliantha subsp. bryophila, which is a circular molecule of 152,045 bp in length, including a large single-copy region (83,966 bp), a small single-copy region (17,663 bp), and a pair of inverted repeats (25,208 bp). The chloroplast genome contained 113 genes, including 79 protein-coding genes, four rRNA genes, and 30 tRNA genes. The phylogenetic tree based on chloroplast genomes showed the relative relationship of P. calliantha subsp. bryophila and P. calliantha, which further supports P. calliantha subsp. bryophila as a subspecies of P. calliantha in taxonomy. The complete chloroplast (cp) genome of P. calliantha subsp. bryophila provides valuable data for further phylogenetic studies of Primulaceae.
Primula calliantha subsp. bryophila is a perennial alpine species in the Sect. Crystallophlomis Rupr. of the family Primulaceae. P. calliantha subsp. bryophila is a subspecies of P. calliantha, the whole chloroplast genome of P. calliantha (NC_034678) and its phylogenetic relationship with related species have been reported (Yang et al. Citation2021). The main difference from the original variant is that the calyx is shorter and the corolla tube is narrower. P. calliantha subsp. bryophila distribute in northwestern Yunnan and adjacent eastern Tibet, growing in alpine grasslands and rhododendron bushes at an altitude of 3800–4500 meters. It is also distributed in northern Myanmar (Hu Q and Kelso Citation1996). In this research, we first report the complete chloroplast genome sequence of P. calliantha subsp. bryophila and constructed a phylogenetic tree, which provided a scientific basis for understanding its taxonomic status.
In this research, the fresh leaves and specimen of P. calliantha subsp. bryophila were collected from Zhongdian Tianchi, Yunnan, China (N 27°37′, E 99°38′). The specimen was deposited in the Herbarium of Yunnan Normal University under the voucher number of HY20 (Kunming, China; Jianlin Hang, [email protected]). Total genomic DNA was extracted using a modified CTAB (cetyl trimethylammonium bromide) method (Sahu et al. Citation2012). The fragmented genomic DNA was used to construct short-insert libraries for Illumina paired-end (PE) sequencing on the Illumina Hiseq X Ten sequencer. The clean reads were assembled by NOVOPlasty v2.7.2, and two alternative chloroplast sequences in opposite directions were obtained (Dierckxsens et al. Citation2016). Then the cp genome was annotated using Geneious v8.0.2 software based on the reference of P. calliantha (MZ054238) (Matthew et al. Citation2012).
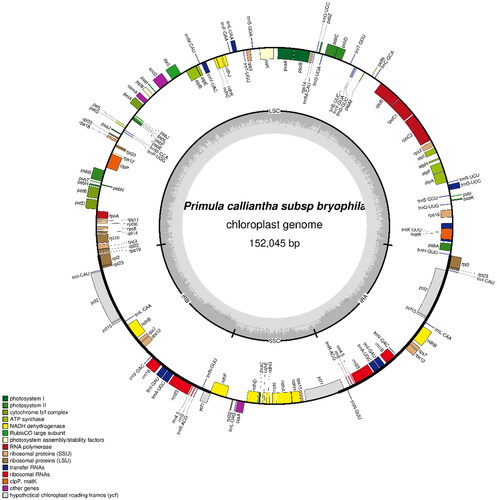
The raw reads (19,100,898) were subjected to assembly to produce a circular molecule of complete chloroplast with about an average 831.8 coverage. The cp genome of P. calliantha subsp. bryophila (GenBank accession ON416873) is 152,045 bp in length, including a large single-copy region (83,966 bp), a small single-copy region (17,663 bp), and a pair of inverted repeats (25,208 bp). The overall GC content is 37%. In total, the cp genome was annotated with 113 genes, including 79 protein-coding genes, 30 tRNA genes and four rRNA genes.
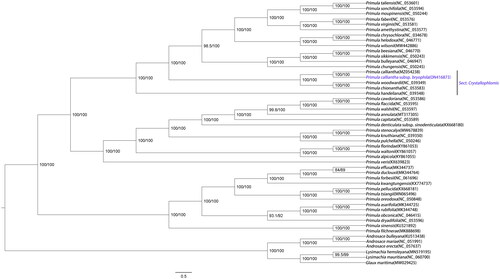
In order to explore the phylogenetic relationship of P. calliantha subsp. bryophila within the genus Primula, 43 Primula cp genomes were downloaded as well as three Androsace species, two Lysimachia species and Glaux maritima (Liu et al. Citation2021) as outgroups from GenBank. These genome sequences were aligned by MAFFT software (Katoh and Standley Citation2013), and then a maximum likelihood phylogenetic tree was constructed using IQ_TREE v1.6.12 (Nguyen et al. Citation2015). The branch support was tested with 10,000 replicates using the SH-like approximate likelihood ratio (SHAlrt) (Guindon et al. Citation2010) and ultrafast bootstrap approximation (UFboot) (Hoang et al. Citation2018). The optimal model according to the Bayesian information criterion was TVM + F+R3 using ModelFinder (Kalyaanamoorthy et al. Citation2017) and used FigTree v1.4.4 software to further beautify the phylogenetic tree (). The phylogenetic tree based on chloroplast genomes showed the relative relationship of Primula calliantha subsp. bryophila and P. calliantha, which further supports P. calliantha subsp. bryophila as a subspecies of P. calliantha in taxonomy (). The complete chloroplast genome of P. calliantha subsp. bryophila provides valuable data for further phylogenetic studies of Primulaceae ().
Figure 1. ML phylogenetic tree of Primula calliantha subsp. bryophila and 49 Primulaceae species based on complete chloroplast genome, branch supports values were reported as SH-aLRT/UFBoot.
Ethics statement
Complying with ethics of experimentation: Plant samples used in this study are unprotected species. We confirm that all research is conducted in accordance with ethical guidelines and the legal requirements of the country.
Author contributions
Rui Li is mainly responsible for the data analysis and writing of the manuscript, Li Zhang is responsible for the guidance and actual participation in the related data analysis. In addition to collecting plant materials and critically revising the manuscript, Yunqi Liu and Shubao Wang also made substantial contributions to the conception of the work. Yuan Huang is responsible for supervising the project and reviewing and approving the final version of the manuscript to be released.
Disclosure statement
The authors declare that they have no competing interests.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov/] under the accession no. ON416873. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA834972, SRR19141994, and SAMN28088249, respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2016. Novoplasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):9.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Hu Q, Kelso S. 1996. Primulaceae. In: Wu CY, Raven PH, editors. Flora of China. Vol. 15. Beijing and St. Louis: Science Press and Missouri Botanical Garden Press; p. 185–188.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. Mafft multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu YQ, Chen X, Zhang L, Huang Y. 2021. The complete chloroplast genome of Glaux maritima, a monotypic species. Mitochondrial DNA B Resour. 6(8):2137–2138.
- Matthew K, Richard M, Amy W, Steven SH, Matthew C, Shane S, Simon B, Alex C, Sidney M, Chris D. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Sahu SK, Thangaraj M, Kathiresan K. 2012. DNA extraction protocol for plants with high levels of secondary metabolites and polysaccharides without using liquid nitrogen and phenol. ISRN Mol Biol. 2012:205049–205049.
- Yang X, Huang Y, Li Z, Chen J. 2021. Complete plastid genome of Primula calliantha Franch. (Primulaceae): an alpine ornamental plant endemic to Hengduan Mountain. Mitochondrial DNA B Resour. 6(9):2643–2645.