Abstract
The Pseudohemiculter hainanensis (Boulenger, 1900) is a small Cyprinidae fish that has a wide distribution in China. In this study, we characterized the complete mitochondrial genome of P. hainanensis by the Illumina NovaSeq sequencing platform in Guangxi, China. The assembled mitogenome is 16,647 base pairs (bp) and consists of 13 protein-coding genes (PCGs), 22 transfer RNAs, two ribosomal RNAs, and a control region (D-loop). Nucleotide composition of the complete mitogenome is 29.69% (A), 24.82% (T), 27.97% (C), and 17.52% (G), with an A + T bias of 54.51%. The maximum-likelihood tree based on 13 PCGs showed that Pseudohemiculter hainanensis formed an independent lineage and P. hainanensis was closer to T. houdemeri.
Introduction
The Pseudohemiculter hainanensis (Boulenger, 1900) is a small Cyprinidae fish, that is widely distributed in the drainage basins of the Yuanjiang River, the Pearl River, Hainan Island, and the middle reaches of the Yangtze River in China. It is a freshwater fish that lives in the upper middle layer of the water (Luo and Chen Citation1998). P. hainanensis was classified as least concern (LC) according to International Union for Conservation of Natures. No research on P. hainanensis has been documented till date; therefore, it is highly important to obtain the complete mitochondrial genome of P. hainanensis for further studies. This study obtained the complete mitochondrial genome of P. hainanensis, analyzed the structural features of the complete mitochondrial genome, and explored the phylogenetic relationships within Cultrinae to provide a basis for further studies on the genetic evolution and classification of P. hainanensis.
Materials
Specimen of P. hainanensis was collected from the farmers market of Laibin, Guangxi Zhuang Autonomous Region, China (23°43′24″N, 109°13′45″E) on 5 July 2021. The species was carefully distinguished as ‘Cypriniformes, Cyprinidae, Cultrinae, and Pseudohemiculter’ according to Culterinae. Fauna Sinica, Osteichthyes, Cypriniformes II book (Luo and Chen Citation1998). The voucher samples were deposited at the college of environmental science and engineering, Guilin University of Technology (Zhiqiang Wu, [email protected], under the voucher number pha202105).
Methods
One P. hainanensis captured in 2021 was dissected using a sterilized dissection tool, and 5 g of its dorsal muscle was taken and stored in a centrifuge tube at −80 °C in the refrigerator. Total genomic DNA was extracted using the cetyltrimethylammonium bromide (CTAB) method (Chapela et al. Citation2007). The extracted DNA was used to construct the library by the Whole Genome Shotgun (WGS) method. Covaris M220 ultrasonic fragmentation machine was used to split the fragments into 400 bp lengths fragments with splices at both ends, and then bridge PCR amplification was performed. The constructed library was sequenced paired-end (PE) using next-generation sequencing (NGS) based on the Illumina NovaSeq sequencing platform. A5-miseq v20150522 (Coil et al. Citation2015) and spadesv3.9.0 were used to de novo assemble high-quality second-generation sequencing data to construct contig and scaffold sequences. The spliced genome sequence was uploaded to the MITOS web (http://mitos.bioinf.uni-leipzig.de/) server for functional annotation (Bernt et al. Citation2013). Among them, the genetic code is set to 02 vertebrate, and the rest are set according to the default parameters set by MITOS.
Results
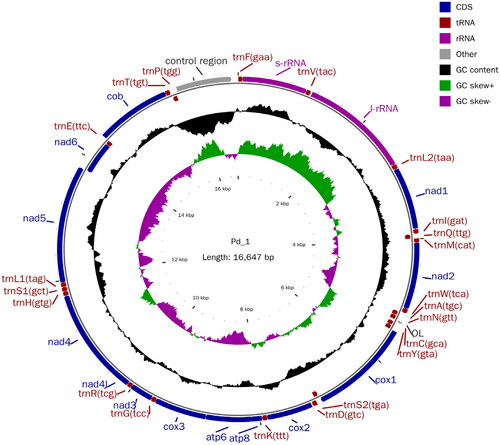
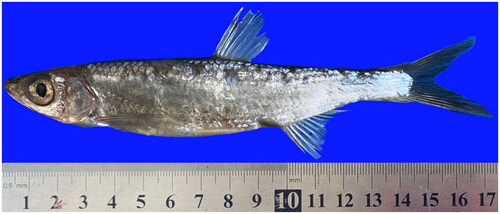
The P. hainanensis’s body length is 5–5.6 times its height and P. hainanensis’s pectoral fin has 15 branching fins with a scaleless keel between ventrals and anal. The photo of the sample was taken by us in the field and is shown in . The complete mitogenome of P. hainanensis was sequenced to be 16,647 bp in length and exhibits an average GC content of 45.49%. The complete mitochondrial genome comprised 13 protein-coding genes (PCGs), 22 transfer RNA genes, two ribosomal RNA, and a control region (D-loop). Most PCGs of P. hainanensis were encoded on the N-strand, except for ND6 which was encoded on the J-strand. The overall base composition was 29.69% (A), 24.82% (T), 27.97% (C), and 17.52% (G), with an A + T bias of 54.51%. Twelve genes started with ATG while only COI started with GTG among the 13 PCGs. Eight genes shared the termination codon TAA including ND2, COI, COII, COIII, ATP6, ND4, ND4L, and Cytb, and the gene with TAG as the stop codon was ND1, ATP8, ND3, ND5, and ND6. The genome map is shown in .
Figure 1. The photo of P. hainanensis (this photo was taken by Yifei Wang, the first author of this article).
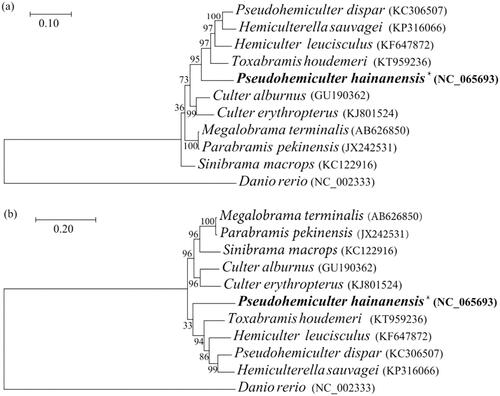
To investigate the phylogenetic relationships of P. hainanensis and other 10 species from Cultrinae, phylogenetic trees were obtained using maximum-likelihood (ML) analyses based on the combined DNA sequences of 13 PCGs and corresponding amino acids (Ai et al. Citation2013; Imoto et al. Citation2013; Xiang et al. Citation2013; Zhang et al. Citation2014; Chen, Wang, et al. Citation2016; Chen, Li, et al. Citation2016; He et al. Citation2016; Wang et al. Citation2017). Danio rerio from Danioninae was selected as an outgroup. ML trees were based on 13 PCGs (), while ML trees were on the basis of amino acids (). The phylogenetic analysis showed that all fishes of the genus Cultrinae were clustered into one group. Both trees showed that P. hainanensis formed an independent lineage and P. hainanensis was closer to T. houdemeri.
Discussion and conclusions
The gene arrangement pattern and translated orientation of P. hainanensis were identical to most vertebrates (Boore Citation1999). The phylogenetic analysis showed that P. hainanensis and Toxabramis houdemeri were in the same proximal branch, and P. hainanensis was differentiated earlier. Dai et al. (Citation2005) used the characteristics of bone differences to study the phylogeny of Cultrinae. They found that genetic differentiation between Pseudohemiculter, Toxabramis, and Hemiculter was small and the genetic relationship was close. All of the above showed that P. hainanensis and T. houdemeri may have similar genetic backgrounds and need further research.
The results of this study showed that P. hainanensis formed an independent lineage and P. hainanensis was closer to T. houdemeri. This study can provide important basic information for species identification, geographic population division, and kinship identification of P. hainanensis in the freshwater waters of China.
Ethical approval
Experiments were performed in accordance with the recommendations of the Ethics Committee of the Guilin University of Technology. These policies were enacted according to the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee protocols.
Author contributions
Yifei Wang: interpretation of the data and writing, revising the draft. Jiayang He: conception, design, and analysis. Zhiqiang Wu and Liangliang Huang: design and the final approval of the version to be published. Minghui Gao, Mingsi Li, and Jie Feng: analysis of the data and identifying species. All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession number NC_065693. The associated BioProject, SRA, and Bio-Sample numbers of specimen are PRJNA832045, SRR18919243, and SAMN27773197, respectively.
Additional information
Funding
References
- Ai W, Chen X, Xiang D, Chen Y, Chen S. 2013. Complete mitochondrial genome of the Sinibrama macrops (Cyprinidae: Cultrinae). Mitochondrial DNA. 24(3):231–233.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. Stadler PF: MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Chapela MJ, Sotelo CG, Pérez-Martín RI, Pardo MÁ, Pérez-Villareal B, Gilardi P, Riese J. 2007. Comparison of DNA extraction methods from muscle of canned tuna for species identification. Food Control. 18(10):1211–1215.
- Chen H, Wang D, Duan X, Liu S, Chen D. 2016. The mitochondrial genome of Pseudolaubuca sinensis (Cypriniformes, Cyprinidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3356–3357.
- Chen L, Li B, Zhou L, Zhao G. 2016. The complete mitochondrial genome sequence of Predatory carp Chanodichthys erythropterus (Cypriniformes: Cyprinidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):1119–1120.
- Coil D, Jospin G, Darling AE. 2015. Darling AE: a 5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589.
- Dai Y, Yang J, Chen Y. 2005. Phylogeny and zoogeography of the subfamily Cultrinae (Cyprinidae). Acta Zootaxon Sin. 30(2):213–233.
- He B, Chen Y, Liu Y, Du J, Deng X. 2016. The complete mitochondrial genome of Hemiculterella sauvagei (Teleostei, Cyprinidae, Hemiculterella). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3322–3324.
- Imoto JM, Saitoh K, Sasaki T, Yonezawa T, Adachi J, Kartavtsev YP, Miya M, Nishida M, Hanzawa N. 2013. Phylogeny and biogeography of highly diverged freshwater fish species (Leuciscinae, Cyprinidae, Teleostei) inferred from mitochondrial genome analysis. Gene. 514(2):112–124.
- Luo Y, Chen Y. 1998. Culterinae. Fauna Sinica, Osteichthyes, Cypriniformes II. Beijing: Science Press.
- Wang P, Chen S, Ou Y, Wen J, Li J. 2017. Complete mitochondrial genome of Toxabramis houdemeri (Cypriniformes; Cyprinidae) and its phylogenetic analysis. Mitochondrial DNA A DNA Mapp Seq Anal. 28(2):292–293.
- Xiang D, Ai W, Chen X, Chen Y, Chen S. 2013. Complete mitogenome of Pseudohemiculter dispar (Cyprinidae: Cultrinae). Mitochondrial DNA. 24(4):356–358.
- Zhang X, Song W, Wang Y, Du R, Wang W. 2014. The complete nucleotide sequence of white Amur bream (Parabramis pekinensis) mitochondrial genome. Mitochondrial DNA. 25(5):361–362.