Abstract
Wallacea dactyliferae Maulik Citation1919 (Coleoptera: Chrysomelidae) has been reported as a new invasive palm pest in Asia recently. So far, a total of 29 species have been reported in Wallacea. In the present study, the whole mitochondrial genome of W. dactyliferae was identified for the first time (also for the first species of Wallacea) by using high throughput sequencing systems. The entire genome is 16,243 bp in length (ACCN: OK513040) consisting of 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and an A + T-rich region. Phylogenetic analysis revealed that insects from the same subfamily were clustered together, with W. dactyliferae being clustered together with other Cassidinae species. This study can provide essential DNA molecular data for further phylogenetic and evolutionary analyses for Chrysomelidae family of the Coleoptera order.
So far, a total of 29 species have been reported in Wallacea (Coleoptera: Chrysomelidae) (Sekerka Citation2015, Lee and Sekerka Citation2018), all distributed in the Oriental Region. Wallacea dactyliferae was first reported to feed on date palm Phoenix dactylifera in India (Maulik Citation1919) and has become a serious pest on P. canariensis, Chamaerops humilis and Washingtonia filifera in France (Drescher and Martinez Citation2005) following accidental introduction. Wallacea dactyliferae has been reported as a new invasive palm pest in Asia recently. It originated in southern India and then spread to Bangladesh, Vietnam, Thailand, Taiwan and Yunnan of China (Sekerka Citation2015; Lee and Sekerka Citation2018; Wu Citation2019). This pest has a certain ability to invade a new environment, and may further spread the danger in China in the future.
In this study, W. dactyliferae was collected in 2021 in Anning (Latitude: 24.9461 N; Longitude: 102.4914E) of Yunnan province, China and the specimen (voucher no. M2021-0127; ) was deposited in the Insect Systematics and Diversity Lab at Yunnan Academy of Biodiversity, Southwest Forestry University, Kunming, China (https://yab.swfu.edu.cn/; contact person, Song Yang, email: [email protected]). Genomic DNA was extracted from the whole body of a single W. dactyliferae pupa using the CTAB method (Doyle and Doyle Citation1987). The isolated DNA was sheared to 300-500 bp fragments in a Covaris (KBiosciences) ultrasonicator device. The sequencing cDNA libraries were prepared by using TruSeq Nano DNA Sample Prep Kit (Illumina, San Diego, CA) following the manufacturer’s protocols. The library preparations were sequenced on an Illumina NovaSeq 6000 platform and 150 bp paired-end reads were generated. The clean reads were generated from the raw reads by trimming adapters and low quality reads by using Trimmomatic v0.39 (http://www.usadellab.org/cms/index.php?page=trimmomatic). The mitochondrial genome of W. dactyliferae was assembled by using NOVOPlasty (Dierckxsens et al. Citation2017) and genes within the genome were predicted by DOGMA (Wyman et al. Citation2004) and MITOS (Bernt et al. Citation2013).
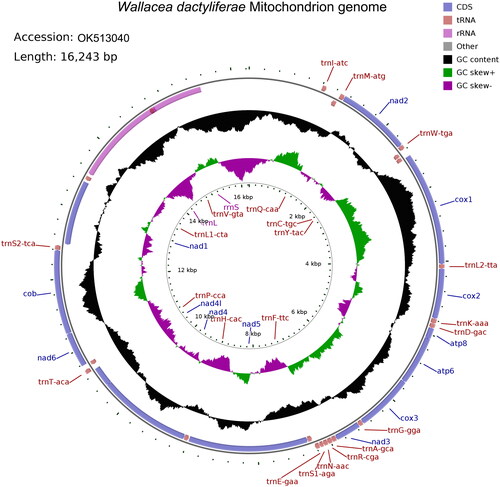
The W. dactyliferae mitochondrial genome (GenBank ID: OK513040) was 16,243 bp in size, circular and double-stranded, with GC content being 23.01% (). This genome has 2 rRNAs, 22 tRNAs and 13 protein-coding genes (PCGs), and a non-coding A + T-rich region (1696 bp). The minority strand (N-strand) of the genome encodes 14 genes, including 2 rRNAs, 8 tRNAs and 4 PCGs; and the majority strand (J-strand) encodes 23 genes, including 14 tRNAs and 9 PCGs. Three kinds of start codons are found in PCGs, namely, ATT, ATA and ATG. Five PCGs end with TAA and one PCG ends with TAG, and the other PCGs have incomplete stop codons, with one being TA- and six being T–. The order and orientation of the above mitochondrial genes, as well as the start and end codons used by these genes were similar to other Cassidinae insects (Xu et al. Citation2018; Yan et al. Citation2021; Zhang et al. Citation2021).
Figure 2. Mitochondrial genome pattern map of W. dactyliferae. The assembled mitochondrial genome (16,243 bp) of W. dactyliferae (GenBank ID: OK513040) with major features: there are 13 protein-coding genes, 22 tRNA genes, the 12S and 16S rRNA genes; different colors represent different gene blocks, with the arrow pointing in the transcription directions. This map was drawn using the online mitochondrial visualization tool CGView (http://stothard.afns.ualberta.ca/cgview_server/).
The complete mitochondrial genomes of W. dactyliferae and other Coleoptera species from Genbank were downloaded and aligned with the MAFFT program (Katoh et al. Citation2005). The phylogenetic relationship of these species were analyzed by using MEGA 7.0 software based on maximum-likelihood (ML) method with Poisson correction model and 1,000 bootstrap replicates (Tamura et al. Citation2013). Phylogenetic analysis revealed that species from the same subfamily were clustered together (), with W. dactyliferae being clustered together with other Cassidinae species. The phylogram is consistent with the taxonomic classification of insects.
Figure 3. The maximum likelihood (ML) phylogenetic tree of W. dactyliferae and other Coleoptera species. Two Lampyrinae species were used as outgroups. Support values next to the nodes are based on 1000 bootstrap replicates.

In a word, this study identifies the entire mitochondrial genome of W. dactyliferae for the first time, which can provide necessary DNA molecular data for phylogenetic analyses of the Chrysomelidae.
Ethical permissions
Insects for which a permit is not required.
Acknowledgements
We are grateful to Dr. Jin Xun for the revision of this article and Mr. Xinxin Xiong for the assistance of insect collection and photographing.
Authors’ contributions
S.Y., G.Y. and Z.X. collected the insects; S.Y., G.Y. and J.H. designed the study; S.Y., G.Y. and Z.X. analyzed the data; S.Y., G.Y. and J.H. wrote the paper. All authors read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitochondrial genome data supporting this study are openly available in GenBank at: https://www.ncbi.nlm.nih.gov/nuccore/OK513040. Associated BioProject, SRA, and BioSample accession numbers are https://www.ncbi.nlm.nih.gov/bioproject/ PRJNA854766, https://www.ncbi.nlm.nih.gov/sra/SRR20706871, and SAMN29446782, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle J, Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissues. Phytochem. Bull. 19:11–15.
- Drescher J, Martinez M. 2005. Le coléoptère Pistosia dactyliferae menace les palmiers du sud de la France. PHM – Reuve Hortic. 448:34–35.
- Katoh K, Kuma K-I, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33(2):511–518.
- Lee C-F, Sekerka L. 2018. Taxonomic status of Wallacea dactyliferae Maulik (Coleoptera: Chrysomelidae: Cassidinae: Bothryonopini) from Taiwan. Jap J Syst Entomol. 24:299–303.
- Maulik S. 1919. Coleoptera. Chrysomelidae (Cassidinae and Hispinae). In: Shipleys AE, editor, The Fauna of British India, including Ceylon and Burma. London: Taylor and Francis; p. 439.
- Sekerka L. 2015. Wallacea, Pistosia and Neodownesia: three distinct genera and their tribal placement (Coleoptera: Chrysomelidae: Cassidinae). Acta Entomol Mus Nat Prag. 55:713–743.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Wu J-L. 2019. Adaptability to environmental temperature of invasive insect pest Wallacea dactyliferae on the palmaes. In: College of plant protection. Fujian: Fujian Agriculture and Forestry University; p. 75.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Xu JS, Liao CQ, Guo QY, Long CP, Dai XH. 2018. Mitochondrial genome of a leaf-mining beetle Prionispa champaka Maulik (Coleoptera: Chrysomelidae: Cassidinae). Mitochondrial DNA B Resour. 3(1):147–148.
- Yan SQ, Lyu B, Tang X, Lu H, Tang JH, Meng R, Cai B, Yang F. 2021. Complete mitochondrial genome of nipa palm hispid beetle Octodonta nipae Maulik (Coleoptera: Chrysomelidae: cassidinae). Mitochondrial DNA B Resour. 6(9):2652–2653.
- Zhang SD, Guo QY, Xu JS, Wang XX, Dai XH. 2021. The complete mitochondrial genome of Downesia tarsata (Coleoptera: Chrysomelidae: Cassidinae). Mitochondrial DNA B Resour. 6(3):1073–1074.

