Abstract
The complete mitochondrial DNA genome of Devario interruptus was sequenced on the Illumina HiSeq platform and found to be 16,735 bp and included 37 genes encoding 13 proteins, 22 tRNAs, two rRNAs, and two non-coding regions. The proportion of nucleotides in mitochondrial genome was T (27.9%), C (23.7%), A (33%), G (15.4%), and the deviation of AT was 60.9%. A Maximum-Likelihood phylogenetic tree was reconstructed using the concatenated mitochondrial protein-coding genes of D. interruptus and other 18 species of fishes. Phylogenetic analysis results supported that D. interruptus was closely related to Devario shanensis. Fundamental genetic data of D. interruptus will be essential for further genetic studies.
1. Introduction
Devario interruptus (Day, 1870), belonging to the subfamily Danioninae, family Danionidae, lives in the slow flow of mountain streams or ditches, and is distributed in Southeast Asian countries on both sides of the Irrawaddy River, especially on the north of Myanmar, and partially distributed in the Nujiang river system, Longchuan Jiang and Daying Jiang watersheds in Yunnan, China (Chu and Chen Citation1989). Barilius interrupta, Brachydanio interrupta, Barilius interruptus, and Danio interruptus are all formerly used taxonomic synonyms for Devario interruptus. The total length of D. interruptus ranges from 42 to 69 mm, and the species is characterized by dorsal fin 3,7; anal fin 2,10–12; pectoral fins 1,10–11; abdominal fin 1,6; longitudinal scales 34–36; transverse scales 9–10; dorsal fin anterior scale 16–18; gill rake 8–9; the head length is smaller than body height; the snout short with a depression in the center of the leading edge; eyes are more proximal to the rostral than to the posterior edge of the gill cover; the mandible is slightly prominent over the maxilla; the mouth cleft reaches up to the nares vertically inferiorly; small round scales cover the body and easy to fall off; the lateral line is incomplete; dorsal fins are free of rigid spines; 4–9 plaques on the body side from the posterior edge of the gill cap to the base of the dorsal fin, then a golden yellow longitudinal band extending to the base of the caudal fin; the ventral side of the male fish is slightly red, while the female fish is slightly yellow (Chu and Chen Citation1989; Fang Citation2000). The reference image was taken by Xiao Jiang CHEN on Jun 10, 2021 ().
Figure 1. Specimen of Devario interruptus was collected from the Yingjiang County, Yunnan Province, China. Photograph by Xiao Jiang CHEN on Jun 10, 2021.

So far, research on D. interruptus is limited to morphological classification (Fang Citation2000; Fang Citation2003), and the lack of genetic information limits our current understanding of the evolutionary phylogenetic relationship of this species within family Danionidae. Therefore, it is of great significance to sequence the whole mitochondrial genome of D. interruptus as soon as possible. This project used high-throughput sequencing technology to carry out sequencing of the whole mitochondrial genome of D. interruptus.
2. Materials and methods
2.1. Sample collection and preservation
In this study, specimens of D. interruptus were collected from the Yingjiang County, Yunnan Province, China (24.493973° N, 97.737320° E) in Jun 2021, using small set nets and gill nets with the permit by Jiangsu Agri-animal Husbandry Vocational College (granted # NSF2021ZR14). To euthanize the fish, the experimental fish were euthanized using eugenol (0.2ml/l), and then transferred to 75% ethanol for 24 hours before being transitioned to 95% ethanol for prolonged storage. All specimens were deposited in the Aquatic Science and Technology Institution Herbarium (https://www.jsahvc.edu.cn/; Voucher number ASTIH-21b0616d02, Chen Xiao Jiang, [email protected]). According to the morphological characteristics of D. interruptus described by Chu and Chen Citation1989, the fish were observed, counted, and measured by using an anatomical microscope and other tools, and finally, the identification was completed.
2.2. DNA extraction, sequencing, and assembly
The tissue sample used for sequencing was kept together with the corresponding voucher samples, the Tguide Cell/tissue genomic DNA Extraction Kit (Tiangen, Beijing, China) was used to isolate DNA from the muscle tissue of a single adult specimen. The concentration of the DNA sample detected by NanoDrop 2000 (Thermo Fisher Scientific, USA). The sequencing library was prepared by random fragmentation of the DNA sample, followed by PCR amplification, size selection, and library quality check. The DNA raw reads were obtained from the sequencing of the constructed library on Illumina HiSeq 4000 Sequencing platform (Illumina, CA, USA). The quality check process was conducted on FastQC 0.11.8 (Andrews Citation2010), the main references are as follows conditions: (1) Sequences containing more than 3 N bases were eliminated; (2) High-quality bases (Phred score ≥20) accounting for less than 60% of sequences were removed; (3) Excluding 3′ end low-quality bases; (4) Sequences less than 60 bp in length were discarded. The sequences were assembled into contigs using metaSPAdes 3.13 (Nurk et al. Citation2017) with default parameters, and Betadevario ramachandrani MH817023 was take as reference (Norén & Kullander Citation2018), and then the resulting D. interruptus draft mitogenome assembly was further analyzed by comparison to the mitogenome of B. ramachandrani (Norén & Kullander Citation2018) both to confirm correct direction assembly of the contigs and identity the starting base position of the D. interruptus mitogenome.
2.3. Annotation and analysis
The resulting circular contig consensus sequence was annotated and verified by mitoMaker 1.14 and MITOS WebServer (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013). Genome Maps was generated using CGView Server (Grant and Stothard Citation2008; https://proksee.ca/). The alignments, analyses, model calculation, and phylogeny reconstruction were all completed by MEGA X (Kumar et al. Citation2018).
3. Results and discussion
3.1. Genomic characterization
The complete mitogenome of D. interruptus was assembled to be a circular DNA molecule of 16,735 bp (T 27.9%, C 23.7%, A 33%, and G 15.4%; 60.9% AT content), that contained 13 protein-coding genes, 22 tRNAs, two rRNAs, and two non-coding regions (OL: origin of L-strand replication, 31 bp; D-loop: displacement loop region, 1125 bp). Among the 37 genes, 9 genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser(UGA), ND6, tRNA-Glu, and tRNA-Pro) were encoded on the L-strand, the other 28 genes were encoded on H-strand. The gene composition, order, and direction were similar to the mitogenomes of species from same subfamily of Danioninae (Song et al. Citation2022). Characteristics of the mitochondrial genome of Devario interruptus was shown in and . The conserved 13 PCGs varied in length from 165 bp (ATP8) to 1,806 bp (ND5). Twelve PCGs have an ATG starting codon except for COX1 that have a GTG starting codon. TAA, TAG, T, and TA were used as the termination codon. The length of 22 tRNAs ranges from 66 bp (tRNA-Cys) to 74 bp (tRNA-Leu). The length of 12S rRNA and 16S rRNA is 954 and 1,651 bp, respectively.
Figure 2. Complete mitochondrial genome map of Devario interruptus (GenBank: MZ853154), with 13 protein coding genes, 22 tRNAs, 2 rRNAs, and 2 non-coding regions. Genes encoded on light strand and heavy-strand were shown inner and outside of the gray circle respectively.
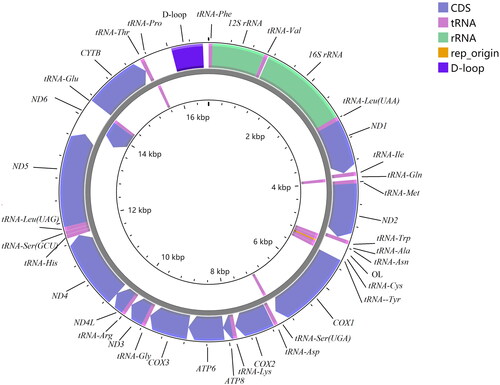
Table 1. Characteristics of the mitochondrial genome of Devario interruptus.
3.2. Phylogenetic analysis
To clarify the phylogenetic position of D. interruptus in subfamily Danioninae, the molecular phylogenetic tree was conducted by the Maximum-likelihood (ML) method based on 13 PCGs of D. interruptus and other 18 published Danionidae species from 8 genera (Devario, Microrasbora, Betadevario, Microdevario, Danio, Barilius, Opsarius, Danionella) (Broughton et al. Citation2001; Tang et al. Citation2010; Lavoué et al. Citation2012; Huang et al. Citation2016; Zhang et al. Citation2016; Zhang et al. Citation2016; Hirt et al. Citation2017; Norén & Kullander Citation2018; Kundu et al. Citation2019; Chen et al. Citation2022; Song et al. Citation2022), and mtREV24 + G + F (the lowest Bayesian information standard score) was selected as the optimal evolutionary model. The result of phylogenetic analysis was summarized in . Six species of the genus Danio were monophyletic, and a sister group to a clade that included D. interruptus, D. kakhienensis, D. devario, M. rubescens, B. ramachandrani, D. laoensis, M. kubotai, and M. nana, and the above 15 species formed a sister group with the genus Danionella.
Figure 3. Phylogenetic reconstruction of D. interruptus and other 18 species based on the concatenated mitochondrial protein-coding genes using Maximum-likelihood method. Barilius bendelisis and Opsarius pulchellus were set to be outgroups. Numbers near the nodes indicated bootstrap support values from 1000 replicates.
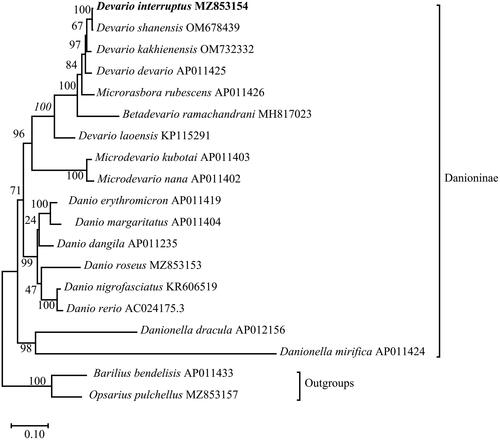
The topology revealed a close relationship between D. interruptus and D. shanensis, and Devario was non-monophyletic, which was inconsistent with Fang’s view that Devario was monophyletic, and Danio did not constitute a sister group with Devario (Fang Citation2003). However, the phylogenetic analysis of this current study agrees with the conclusions of Norén & Kullander (Citation2018), that the genera Devario, Microrasbora, Betadevario, and Microdevario together constitute the sister group of Danio, and support the view of Tang et al. (Citation2010) that Danionella was a sister group of Danio.
4. Conclusion
The complete mitochondrial genome of Devario interruptus was sequenced on the Illumina HiSeq platform to generate a 16,735 bp mitogenome (Genbank accession no. MZ853154). The phylogenetic position of D. interruptus within the subfamily of Danioninae was determined, and the results showed that D. interruptus was closely related to D. shanensis. The mitochondrial genomic data of D. interruptus will be essential for further genetic studies such as evolution, taxonomy, DNA barcoding, resource conservation, and phylogenetic research.
Ethical approval
The experiments were approved by the Ethics Committee for Animal Experiments of Jiangsu Agri-animal Husbandry Vocational College, and conducted following the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Author contributions
Xiao Jiang Chen & Wen Zhao Liu make substantial contributions to the conception or design of the work, and drafting the paper, and Final approval of the version to be published; Lin Song & Hai Xia Liu were involved in the acquisition, analysis and interpretation of the data; the drafting of the paper, and the final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Acknowledgment
The authors acknowledge the Shanghai Genesky Biotechnologies Inc., for their technical support. We thank the anonymous reviewers and editors for their valuable revision comments.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ853154. The associated "BioProject", "Bio-Sample" and "SRA" numbers are PRJNA769978, SAMN22187307, SRR16301567, respectively.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Cambridge, UK: Babraham Bioinformatics, Babraham Institute.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Broughton RE, Milam JE, Roe BA. 2001. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 11(11):1958–1967.
- Chen XJ, Song L, Liu WZ, Gao P. 2022. Characterization of the complete mitochondrial genome and phylogenetic analysis of Opsarius pulchellus (Cypriniformes, Danionidae, Chedrinae). Mitochondrial DNA B. 7(4):666–668.
- Chu XL, Chen YR. 1989. Ichthyology of Yunnan. Beijing (China): Science Press; p. 19–23 (Chinese).
- Fang F. 2000. Barred Danio species from the Irrawaddy River drainage (Teleostei, Cyprinidae). Ichthyological Research. 47(1):13–26.
- Fang F. 2003. Phylogenetic analysis of the Asian cyprinid genus Danio (Teleostei, Cyprinidae). Copeia. 2003(4):714–728.
- Grant JR, Stothard P. 2008. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36(Web Server issue):W181–W184.
- Hirt MV, Arratia G, Chen WJ, Mayden RL, Tang KL, Wood RM, Simons AM. 2017. Effects of gene choice, base composition and rate heterogeneity on inference and estimates of divergence times in cypriniform fishes. Biol J Linnean Soc. 121(2):319–339.
- Huang Q, Ji X, Wang K. 2016. The complete mitochondrial genome of dwarf danio, Danio nigrofasciatus. Mitochondrial DNA A. 27(4):2854–2855.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Kundu S, Chandra K, Tyagi K, Pakrashi A, Kumar V. 2019. DNA barcoding of freshwater fishes from Brahmaputra River in Eastern Himalaya biodiversity hotspot. Mitochondrial DNA B. 4(2):2411–2419.
- Lavoué S, Miya M, Moritz T, Nishida M. 2012. A molecular timescale for the evolution of the African freshwater fish family Kneriidae (Teleostei: Gonorynchiformes). Ichthyol Res. 59(2):104–112.
- Norén M, Kullander S. 2018. The enigmatic Betadevario ramachandrani (Teleostei: Cyprinidae: Danioninae): phylogenetic position resolved by mitogenome analysis, with remarks on the prevalence of chimeric mitogenomes in GenBank. Cogent Biol. 4(1):1525857.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. MetaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834. doi:10.1101/gr.213959.116.
- Song L, Chen XJ, Han XP, Gu YW. 2022. Complete mitochondrial genome and phylogenetic analysis of Danio roseus (Teleostei, Cypriniformes, Danionidae). Mitochondrial DNA B. 7(2):350–352.
- Tang KL, Agnew MK, Hirt MV, Sado T, Schneider LM, Freyhof J, Sulaiman Z, Swartz E, Vidthayanon C, Miya M, et al. 2010. Systematics of the subfamily Danioninae (Teleostei: Cypriniformes: Cyprinidae). Mol Phylogenet Evol. 57(1):189–214.
- Zhang S, Jiang Y, Cui J, Mahboob SK, Ghanim K, Xu P, Sun J. 2016. The complete mitochondrial genome of Microdevario kubotai. Mitochondrial DNA A. 27(2):1150–1151.
- Zhang J, Zhou C, Fei W, Ma L, Kong X. 2016. Complete mitochondrial genome of Danio myersi (Teleostei: Cypriniformes: Cyprinidae). Mitochondrial DNA A. 27(6):3913–3914.
